Fluorescent probe for monitoring apoptosis as well as preparation method and application of fluorescent probe
A fluorescent probe and apoptosis technology, which are applied in the field of fluorescent probes for monitoring apoptosis and their preparation, and can solve problems such as difficulty in detecting apoptosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
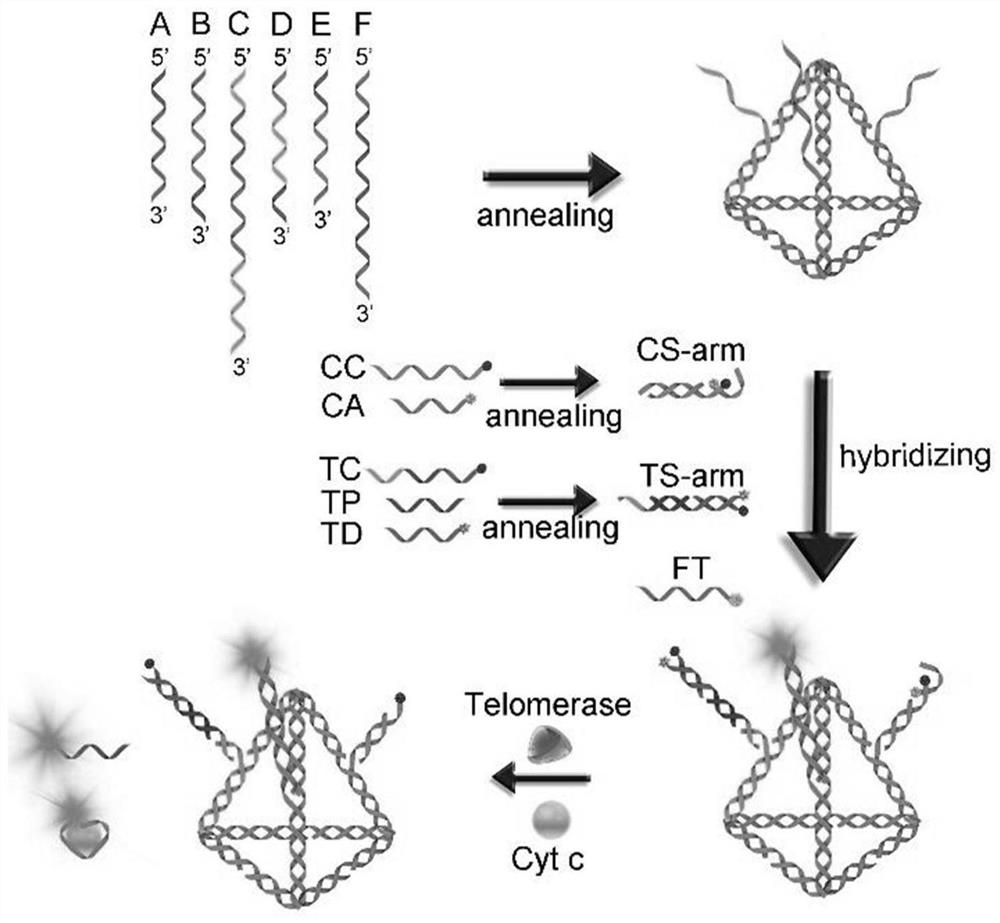
[0094] The preparation process of a fluorescent probe for monitoring the whole process of apoptosis of DNA tetrahedral nucleic acid framework is as follows:
[0095] S1. Preparation of DNA tetrahedral nucleic acid framework:
[0096] Mix equal amounts of ssDNA A, ssDNA B, ssDNA C, ssDNA D, ssDNA E, and ssDNAF in 2×PBS buffer, anneal at 95°C for 10 minutes, and then cool at 37°C for 1 hour to obtain the DNA tetrahedral nucleic acid framework structure.
[0097] S2. Preparation of double-stranded probes:
[0098] 1) Cytochrome c probe: Mix equal amounts of cytochrome c capture chain and cytochrome c aptamer in 2×PBS buffer, anneal at 95°C for 10 minutes, and finally cool at 37°C for 1 hour.
[0099] 2) Telomerase probe: Mix equal amounts of telomerase capture strand, telomerase template strand and telomerase dye strand in 2×PBS buffer, anneal at 95°C for 10 minutes, and finally cool at 37°C for 1 hour.
[0100] S3. Cell apoptosis probe assembly:
[0101] Mix the same amount o...
Embodiment 2
[0102] Example 2 DNA Tetrahedral Nucleic Acid Framework Assembly Process and Reaction Mechanism Characterization
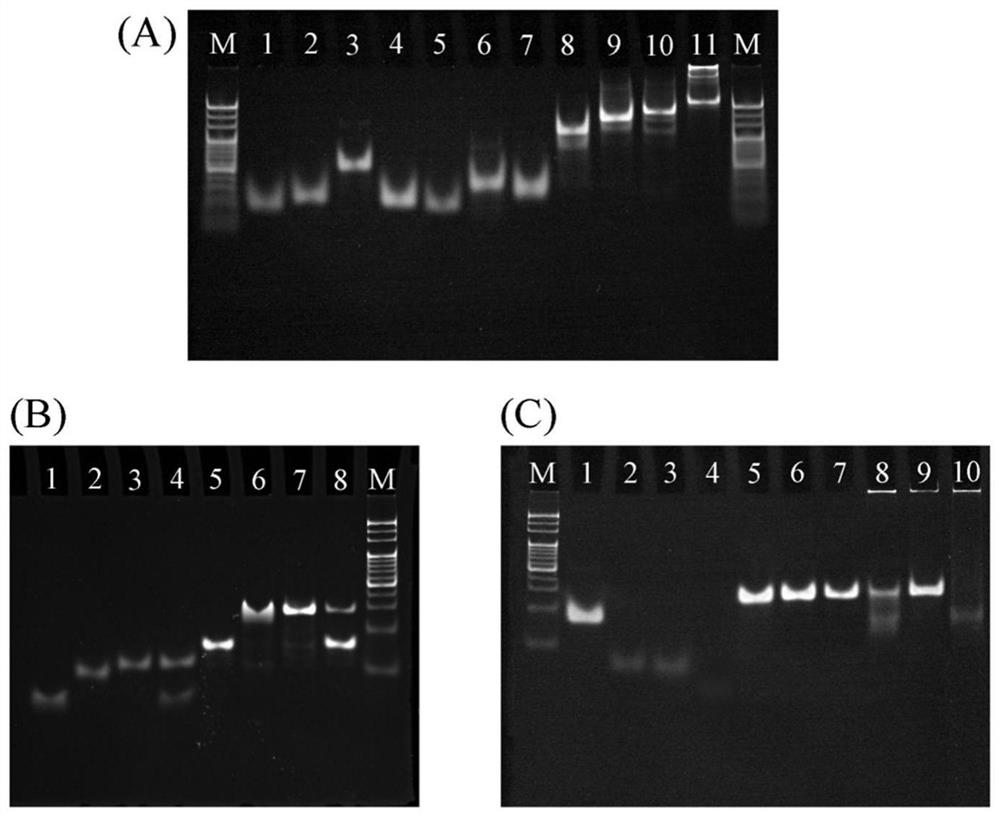
[0103] How the DNA tetrahedron works figure 1 As shown, in order to verify the correctness of the mechanism, use PAGE gel for gel electrophoresis experiment, take 5 μL of DNA sample and mix with 1 μL of 6×DNA loading buffer, run at 80V for 85 minutes and image. PAGE gel electrophoresis to characterize DNA tetrahedron nucleic acid framework-type apoptosis monitoring fluorescent probe construction and its working mechanism.
[0104] In addition to the aforementioned DNA strands, two extended strands of the DNA tetrahedral nucleic acid framework were used for simulating probe binding, the sequences of which are as follows:
[0105] ssDNA C extension:
[0106] ssDNA E extension:
[0107] The formation of the DNA tetrahedral nucleic acid framework was first characterized using 5% PAGE. Such as figure 2 As shown in A, the different bands in lanes 1-6 correspon...
Embodiment 3
[0110] Example 3 Cell Apoptosis Probe Detection of Cell Apoptosis Markers
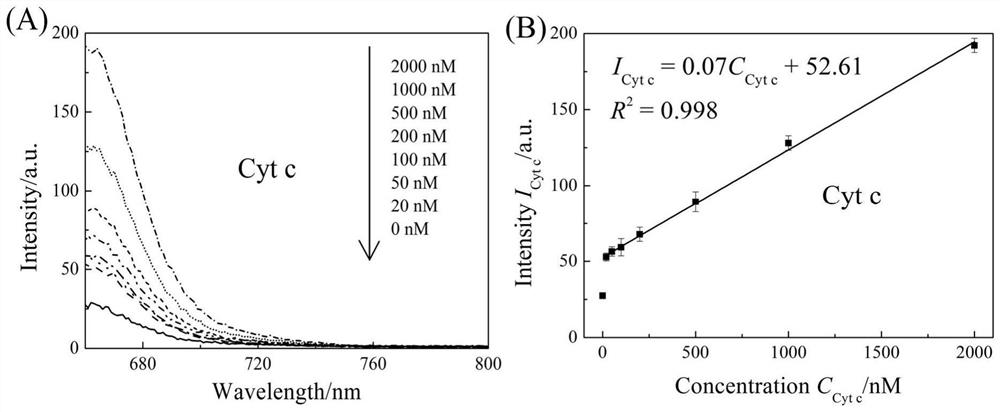
[0111] To the apoptosis probe constructed in Example 1, add cytochrome c or 0, 0.625, 2.5, 5, 12.5, 25, 37.5, 50, 62.5, 70cells / μL of telomerase, the final concentration of the probe is 50nM, and after incubation at 37°C for two hours, the fluorescence intensity is measured using a fluorescence spectrometer, wherein the FAM excitation light is 495nm, and the Cy5 excitation light is 640nm. image 3 Take B and D respectively image 3 Plotting the fluorescence intensity at 660nm and 520nm in A and C.
[0112] image 3 In A, the cytochrome c concentration-dependent Cy5 fluorescence emission line increases with increasing cytochrome c concentration. image 3 B is the linear function of the fluorescence intensity (660nm) of Cy5 as the concentration of cytochrome c changes, that is, I Cyt c =0.07C Cyt c +52.61, R 2 =0.998, LOD=85.93nm, the linear range is 20nm~2000nm. image 3 In C and 3D, the apoptosi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com