Anti-tumor polypeptide, and preparation method and application thereof
An anti-tumor and anti-tumor drug technology, applied in the biological field, can solve the problems of lack of CLDN18.2 research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
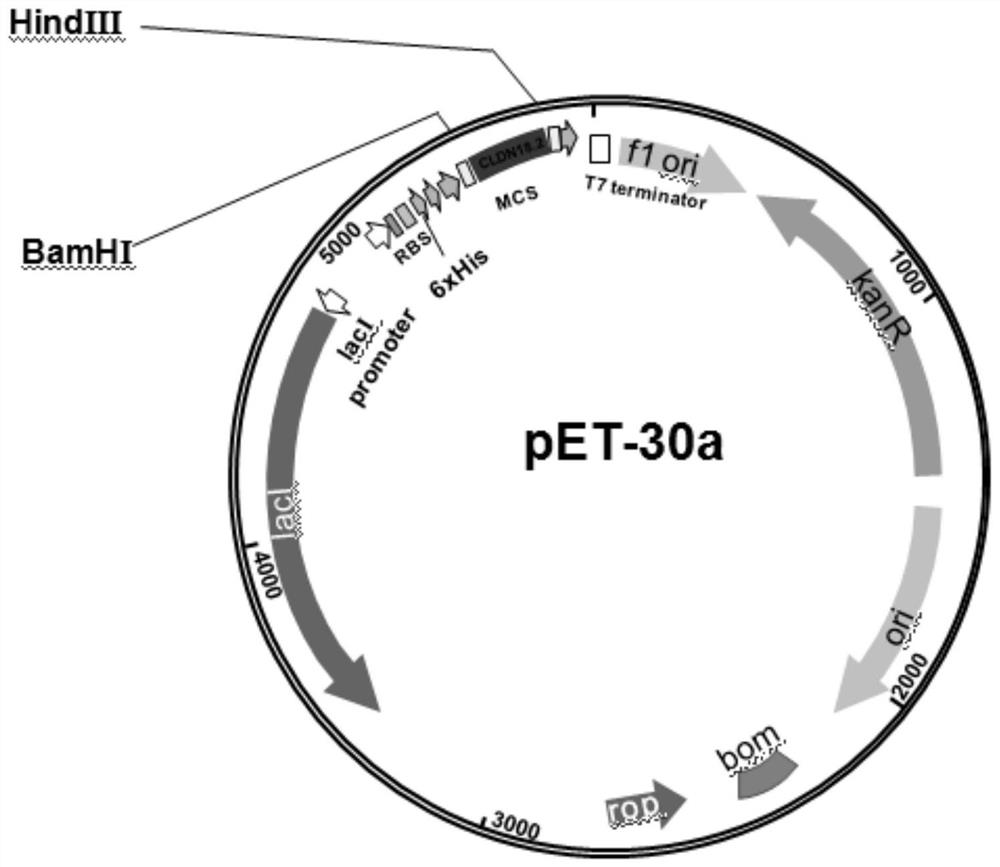
[0018] Example 1: pET-30a / (His) 6 - Construction of CLDN18.2 protein expression vector
[0019] like figure 1 Construct target protein expression vector pET-30a / (His) as indicated 6 -CLDN18.2, search the gene sequence of CLDN18.2 from the Genebank database (1-125 AA, NM_001002026.3), design PCR primers, the upstream primer sequence is: 5'-CG GGATCC ATGGCCGTGACTGCCTGTCAC-3', the downstream primer sequence is: 5'-CC AAGCTT GATGCCTACGATCATCAGGG-3', the underlined part is the restriction site sequence. The PCR product and the vector pET-30a were digested with BamHI and Hind III at 37°C for 3 h, then ligated with T4 DNA ligase at 16°C for 12 h. Transform the ligated product into DH5α competent cells, then spread the transformed product on a kanamycin-resistant (50 μg / ml) LB plate and culture until a single colony grows, pick a single colony, and extract the plasmid for enzyme digestion verification , the recombinant plasmid was sequenced to obtain the recombinant plasmid pET...
Embodiment 2
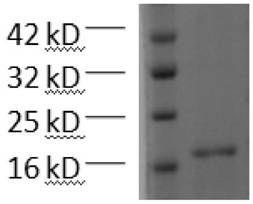
[0020] Example 2: (His) 6 -Expression, purification and validation of CLDN18.2
[0021] The recombinant plasmid pET-30a / (His) prepared in Example 1 6 - CLDN18.2 was transformed into BL21 (DE3) strain, and the recombinant plasmid was screened on a kanamycin-resistant LB plate, and a single colony was picked and cultured in 10 ml LB liquid medium (containing 50 μM kanamycin) until OD 600 After about 0.5, the culture was inoculated in LB liquid medium at a volume ratio of 1: 10, and cultured with vigorous shaking at 37°C until OD 600 About 0.5, add IPTG with a final concentration of 1 mM and induce at 37 °C for 10 h to obtain a bacterial solution.
[0022] Centrifuge the bacterial liquid, remove the supernatant, and separate the bacterial liquid and lysate (50 mM Tris–HCl, 20 mM imidazole, 100 mM NaCl, 10% glycerol, 1% triton, 1 mM protease inhibitor PMSF, 1 mg / ml Lysozyme, pH 8.0) was resuspended in the lysate at a volume ratio of 20:1, placed on ice for 30 min, ultrasonicate...
Embodiment 3
[0024] Example 3. Phage display panning for biologically active peptides that specifically bind to CLDN18.2
[0025] (1) Immobilize the target protein: mix 600 μl of the target protein solution with a concentration of 17 μg / ml (0.1 M NaHCO 3 pH 8.6) into a six-well plate, shake slightly on a shaker, and incubate overnight at 4°C. After washing 6 times with TBST (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% [v / v] Tween-20), the 3 pH 8.6, 5 mg / ml BSA, 0.02% NaN 3 ) closed for 1 h.
[0026] (2) Screening for specifically binding phages: Wash the six-well plate 10 times with TBST. The amplified phage was diluted with TBST to a copy number of 10 9 ~10 11 In between, the diluted phage was added to the six-well plate to allow it to bind to the target protein, and incubated at room temperature for about 60 min. Wash 10 times with TBST and pat dry after each wash. Add eluent to collect phages that specifically bind to the target protein.
[0027](3) Extraction of monoclonal phage...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com