Urinary sediment genome DNA classification method and device and application
A classification method and classification label technology, applied in the fields of genomics and bioinformatics, which can solve the problems of sequencing errors and strong tumor heterogeneity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0171] Embodiment 1: the preparation of DNA sample
[0172] 1. Target group
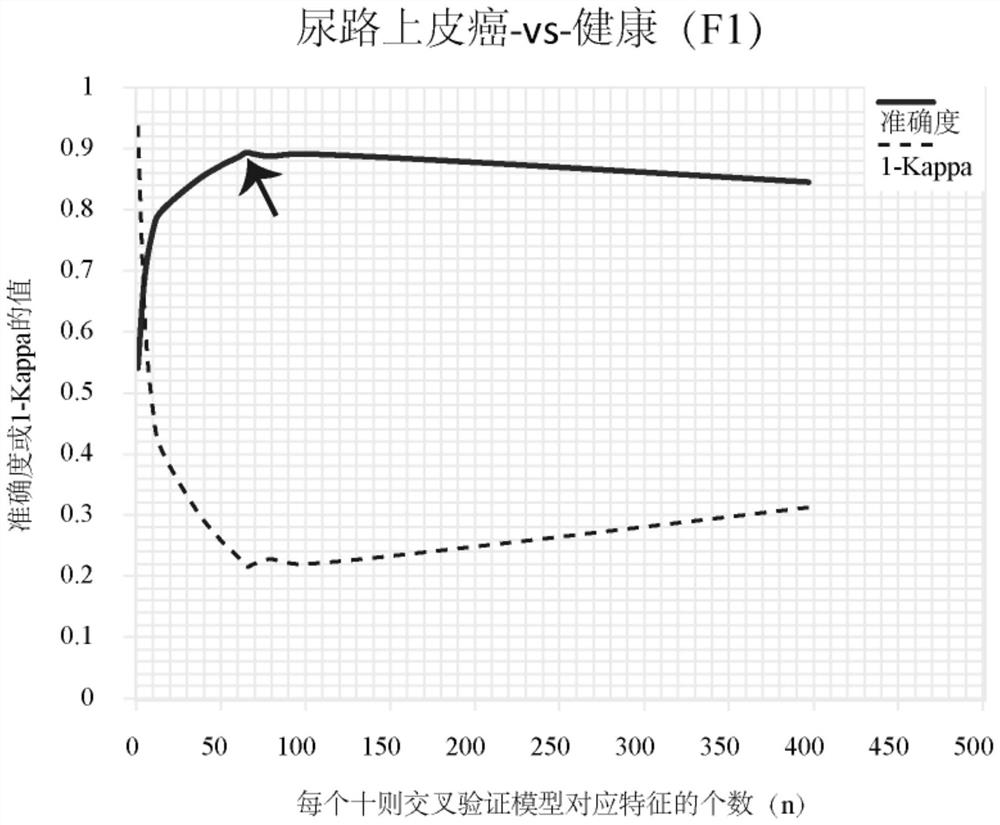
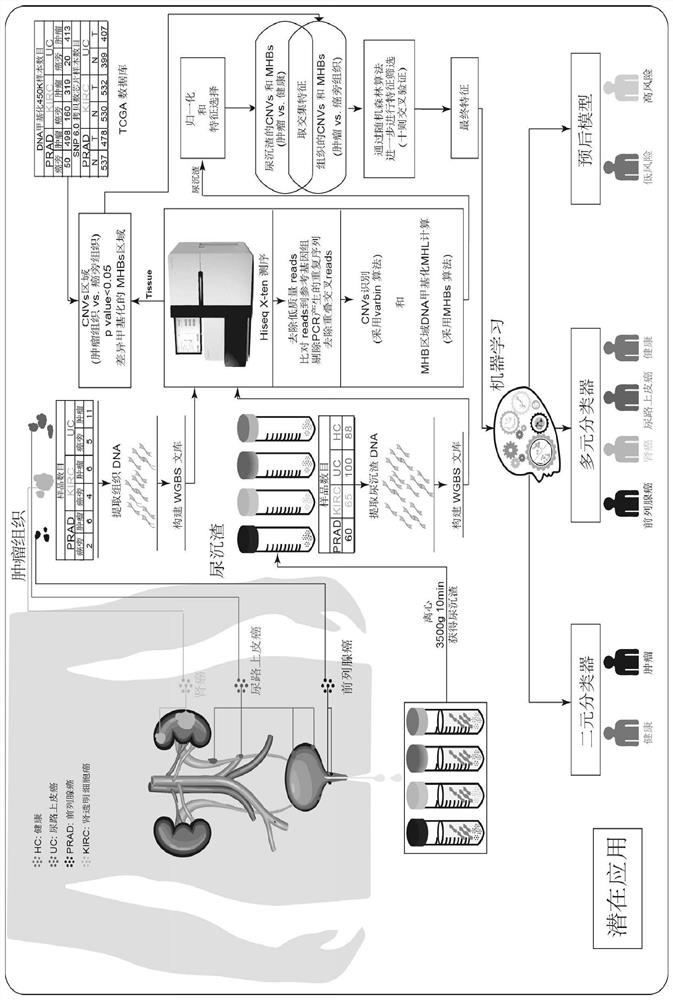
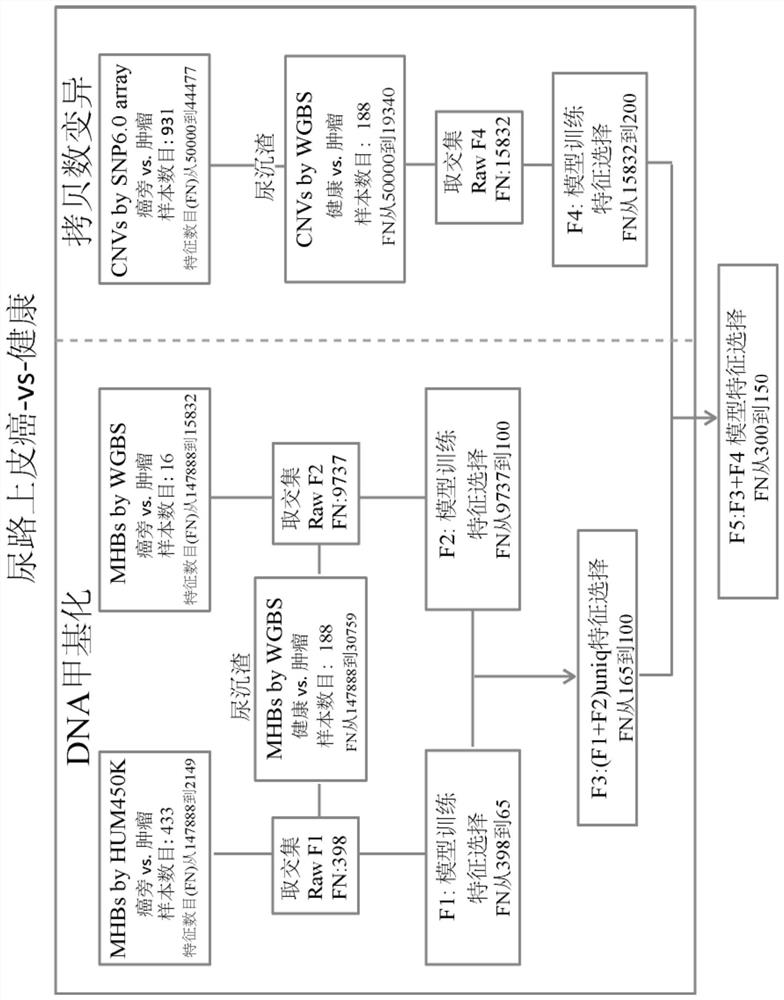
[0173] Urine samples were collected from a total of 313 subjects, such as figure 1 shown. The 313 subjects included 88 healthy people, 65 clear renal cell carcinoma (KIRC) patients, 100 urothelial carcinoma (UC, including bladder cancer UCB, upper tract urothelial carcinoma UTUC) and 60 prostate cancer patients (PRAD) patients.
[0174] 2. Experimental method
[0175] (1) Collect fresh urine (morning urine) from preoperative tumor patients and healthy people (morning urine). The urine is collected in a 50ml centrifuge tube, and the volume of each urine sample is about 45-50ml.
[0176] (2) The collected morning urine samples were centrifuged at 3500 rpm and 4° C. for 10 min, respectively, and the supernatant was removed to obtain urine sediment.
[0177] (3) Wash the urine sediment twice with PBS buffer (add 500ml of PBS buffer each time, centrifuge at 13000g for 1min to remove the supernatant...
Embodiment 2
[0180] Example 2: Whole Genome Bisulfite Sequencing (Whole Genome Bisulfite Sequencing, for short BS-seq or WGBS) library construction
[0181] Take 50-200ng of the DNA samples obtained in Example 1 as the starting DNA for library construction, and add lambda DNA (all CpG sites are unmethylated C) and 5mC DNA (all CpG sites) at a ratio of 3:1000 CpG sites are all methylated C). Then, the DNA was fragmented with a Covaris ultrasonic breaker, so that the main peak of the fragment length was in the range of 400bp. Then use NEBNext Ultra II End Repair / dA-Tailing Module 96rxns (Catalog No. E7546) to repair the end of the fragmented DNA and add polyadenylic acid (polyA), and then use NEBNext Ultra IILigation Module, 96rxns unit (Catalog No. E7595L ) plus a methylated PE linker.
[0182] The obtained water-soluble DNA (library) connected with adapters was treated with bisulfite using the EZ DNA methylhlation Gold kit (Zymo Research), and the specific steps were guided by the ...
Embodiment 3
[0184] Example 3: HiSeq X10 system sequencing
[0185] 1. Sample to be tested:
[0186] The BS-seq library of 313 cases of urinary sediment gDNA prepared in the previous Example 2.
[0187] 2. Experimental method
[0188] Novogene Sequencing Company was commissioned to perform whole-genome sequencing on the BS-seq library of gDNA from 313 cases of urinary sediment.
[0189] 3. Experimental results
[0190]The 150bp pair-end reads (pair-end reads) data of the BS-seq library of 313 cases of urinary sediment gDNA (ie fastq original file) were obtained. Used for subsequent data preprocessing and tumor marker analysis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com