Chroococcidiorella tianjinensis as well as culture method and application thereof
A method for culturing chlorella and a technology for chlorella, which are applied in the field of green algae-Chlorella titanosa and its culturing field, can solve the problems of unknown origin and formation process, unclear genetic background of green algae, errors, etc., so as to improve international competition. strength, good for eye health, clear genetic background
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
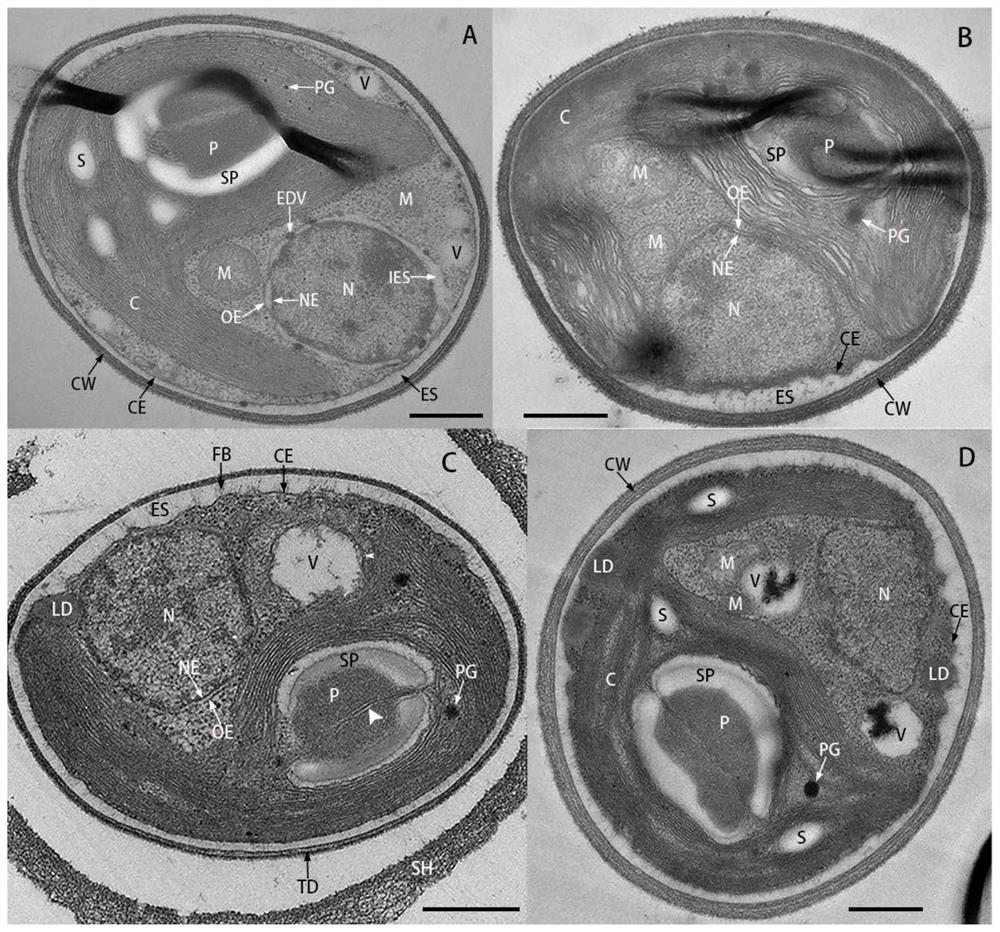
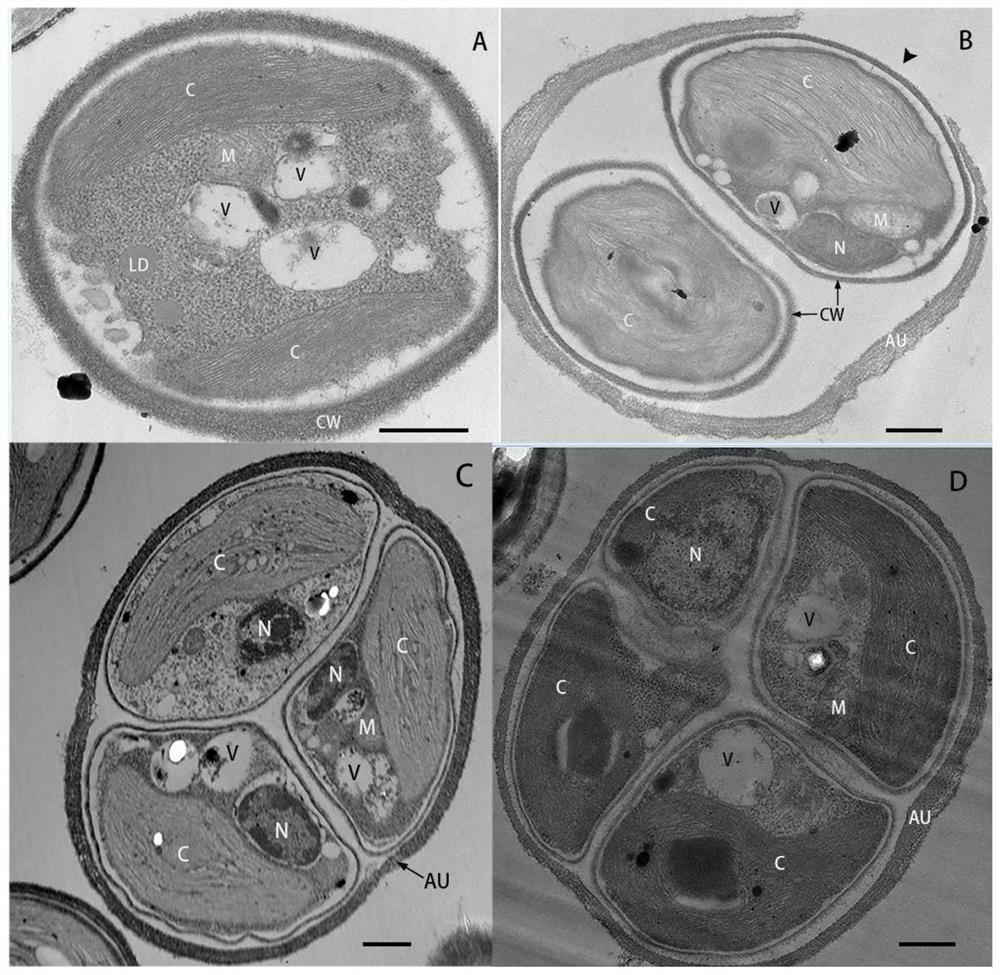
[0041] Example 1 Morphology and ultrastructural observation of TDX16-DE
[0042] 1. Morphological observation of TDX16-DE cells
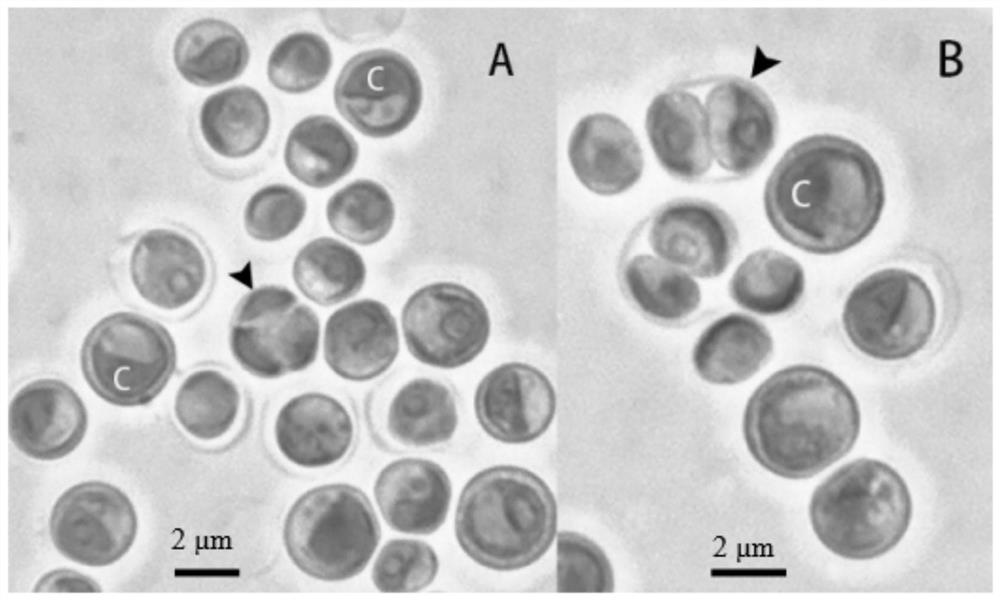
[0043] One inoculation loop of TDX16-DE single colony cells was inserted into 100ml of BBM medium (NaNO 3 , 250mg / l; MgSO 4 .7H 2 O, 75mg / l; NaCl, 25mg / l; K 2 HPO 4 ,75mg / l; KH 2 PO 4 ,175mg / l;CaCl 2 .2H 2 O, 25mg / l; ZnSO 4 .7H 2 O, 8.82 mg / l; MnCl 2 .4H 2 O, 1.44mg / l; MoO 3 ,0.71mg / l;CuSO 4 .5H 2 O, 1.57mg / l; Co(NO 3 ) 2 .6H 2 O, 0.49mg / l; FeSO 4 .7H 2 O, 4.98 mg / l; H 3 BO 3 , 11.4mg / l; KOH, 31mg / l; EDTANa 2 , 50mg / l) in a 200ml conical flask, placed in a light incubator for cultivation, continuous light 60μmol photons m -2 s -1 , 25℃, after culturing for 6 days, take samples to observe the cell morphology and take pictures of the cells under an oil microscope (100×) with a light microscope. like figure 1 As shown, TDX16-DE is green, round or oval, 2.0–3.6 μm in diameter, with 1 adherent chloroplast. Cells proliferate by ...
Embodiment 2
[0048] Example 2 Sequencing of TDX16-DE
[0049] (1) The DNA of TDX16-DE was extracted by CTAB method.
[0050] (2) DNA was amplified with primers RP8 (5'-ACCTGGTTGATCCTGCCAGTAG-3') and RP9 (5'-ACCTTGTTACGACTTCTCCTTCCTC-3') and sequenced. The 18S rRNA sequence of TDX16-DE is shown in SEQ ID NO.1. The blast comparison analysis showed that the 18S rRNA (1687bp) of TDX16-DE had the highest similarity with Chlorella vulgaris, reaching 99.8% (query cover 75%).
[0051] (3) DNA was amplified with primers ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5-TCCTCCGCTTATTGATATGC-3') and sequenced. The ITS sequence of TDX16-DE is shown in SEQ ID NO.2. The results of blast comparison analysis showed that the ITS (761bp) of TDX16-DE had the highest similarity with Chlorella vulgaris, reaching 100% (query cover 99%).
[0052] The 18S rRNA and ITS sequences of TDX16-DE had the highest similarity with Chlorella vulgaris, indicating that TDX16-DE was most similar to Chlorella vulgaris. However, T...
Embodiment 3
[0056] Example 3 Growth of Chlorella tetrazolium Tianjin under different temperature conditions
[0057] An inoculation loop of single algal colony cells of Chlorella tertiaryensis was inserted into a 200ml conical flask containing 100ml of BBM medium, and placed in a light incubator for cultivation at different temperatures, with continuous light of 60μmol photons m -2 s -1 , shake twice a day, after culturing for 10 days, measure the absorbance OD680 of the culture medium. The experimental results showed that Chlorella tianjinensis can grow at 10-40℃, and grow best at 25-35℃.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com