Vaccine fusion protein
A technology of fusion protein and flagellin, applied in the field of fusion protein to achieve the effect of promoting secretion and correct conformation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0175] Example 1 Construction of fusion protein based on flagellin fusion protein
[0176] Construct the fusion protein, from N-terminal to C-terminal: N-terminal constant region of flagellin flagellin (FliC) (amino acids 1-176, SEQ ID NO: 6), B cell epitope hotspot region II (B2, SEQ ID NO: 21, SEQ ID NO: 22 overlaps with SEQ ID NO: 28), SARS-CoV-2 receptor binding domain RBD (contains B1 fragment, SEQ ID NO: 5), T cell epitope (SEQ ID NO: 24 overlaps with SEQ ID NO: 25), panT (pan HLA-DR binding epitope / helper T cell epitope, CD4 + cell epitope, SEQ ID NO: 4), and flagellin FliC C-terminal constant region (amino acids 403-495, SEQ ID NO: 7). Among them, flagellin is an intramolecular adjuvant, which together with the SARS-CoV-2 antigen constitutes a recombinant vaccine, and the obtained fusion protein is called SC2V7 (the amino acid sequence is shown in SEQ ID NO: 9, and the specific structure is shown in figure 2 shown in A).
[0177] Construct the fusion protein, from ...
Embodiment 2
[0180] Example 2 Fusion protein expression verification
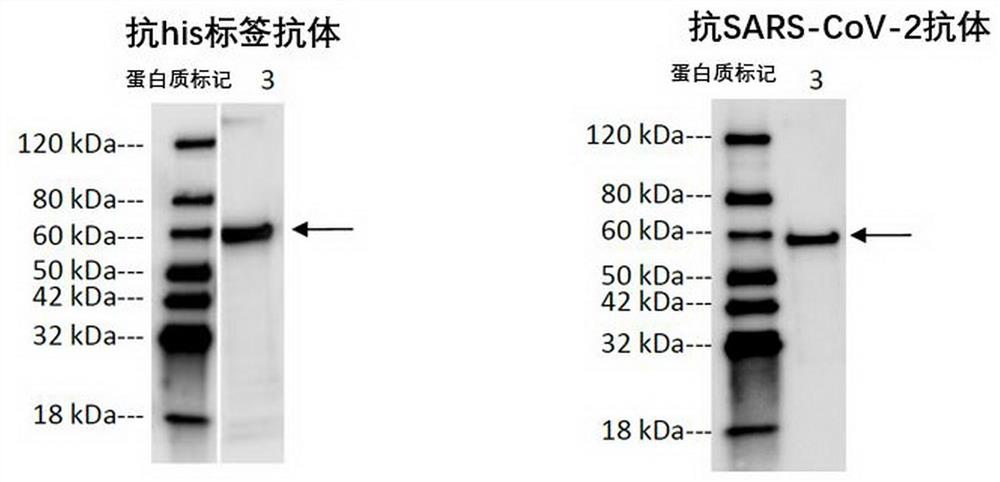
[0181] The nucleic acid sequences encoding SC2V7 and SC2V8 were synthesized by Suzhou Jinweizhi Biotechnology Co., Ltd., and inserted between Nde I and Pst I of the pET21a expression vector (purchased from EMD Biosciences) to obtain an expression vector. The expression vector was transformed into Escherichia coli BL21-Gold (DE3), and cultured overnight at 37°C in LB medium (containing 100 µg / ml ampicillin). The transformed Escherichia coli was expressed on a large scale in TB medium (containing 100 µg / ml ampicillin), and when the DO595 was 2.0-2.5, it was induced by adding 1 mM IPTG (purchased from Sangon Bioengineering (Shanghai) Co., Ltd.), 37°C , and incubated for 4 hours. The cultured bacteria were collected, crushed under high pressure, the bacterial lysate was heated at 42°C overnight, the precipitate was collected by centrifugation, and the precipitate was resuspended in 0.2-0.25M ammonium sulfate solution at 4°...
Embodiment 3
[0183] Example 3 Verification of fusion protein RBD conformation
[0184] 1) The correctly folded RBD domain will be able to interact with its receptor ACE2, and this is the principle to verify the conformation of RBD in recombinant vaccines. The recombinant vaccine protein was obtained according to the method in Example 2. ACE2 protein (Sino biological, Cat: 10108-H08H) was coated on an ELISA microwell plate, and after washing, blocking solution was added to block it, and then the recombinant vaccine protein SC2V7 or SC2V8 was added to the microwell plate. Incubate in the well plate, wash and add anti-RBD antibody (Sino biological, Cat: 40150-T62-COV2) for incubation, wash and then add secondary antibody (Jackson ImmunoResearch, Cat: 111-035-003) for incubation, finally wash and add After the reaction substrate was incubated for an appropriate time, the reaction was terminated, and the data of each well was read to analyze the results. The results showed that both fusion pro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com