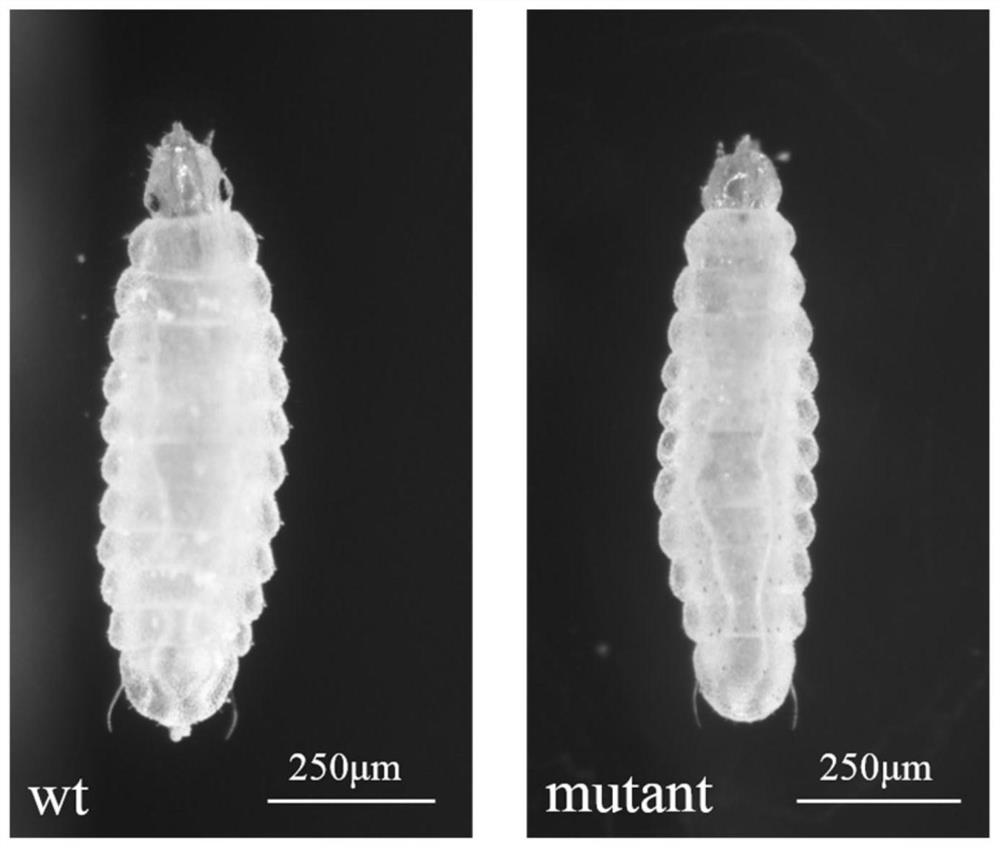
Marker gene of CRISPR/Cas9 gene editing system applied to hermetia illucens
A gene editing and black soldier fly technology, applied in genetic engineering, hydrolytic enzymes, using microinjection methods, etc., can solve the problems of complex mutation screening, achieve excellent results, and facilitate gene function research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The acquisition of embodiment 1 primer
[0031] 1) Extract the total RNA of the black soldier fly in the late stage of embryonic development, reverse transcribe it into cDNA, and clone the white eye color gene white from the black soldier fly cDNA with primers white-F:ATGACTCCAGGCAGCGACGA and white-R:TTAGTTGGCGCCTCTCGCTT, and connect it to P-Jet- 1.2 Sequencing verification on the carrier, the obtained gene sequence is shown as SEQ NO.1 in the sequence listing.
[0032] 2) Select a target site with a length of 23 bases and a PAM sequence of NGG (5'-3') on a single exon of the white gene, and the selected two target site sequences are target site 1 (Target site1:3 'GGCGAACTGCTTGCCATCATGGG 5') and target site 2 (Target site2: 3'GGAGCACCAGGACGTATTAAAGG 5').
[0033] 3) Design primers to synthesize corresponding sgRNA templates, the primer sequences are:
[0034] sgRNA1-F, as shown in SEQ NO.2 in the sequence listing;
[0035] sgRNA2-F, as shown in SEQ NO.3 in the sequen...
Embodiment 2
[0038] Example 2 sgRNA template acquisition
[0039] The template primers sgRNA1-F and sgRNA2-F were respectively annealed and extended with the universal reverse primer to obtain the template (Toyobo).
[0040] The reaction system is as follows:
[0041]
[0042] The PCR reaction conditions are as follows: 94° C. for 2 min; 20 cycles of 94° C. for 15 s, 55° C. for 30 s, and 68° C. for 10 s; 68° C. for 5 min.
[0043] The recovered and purified PCR product is the template sgRNA.
[0044] The sequence of the template sgRNA is:
[0045] sgRNA-1: as shown in SEQ NO.5 in the sequence listing;
[0046] sgRNA-2: as shown in SEQ NO.6 in the sequence listing.
Embodiment 3
[0047] Example 3 A large amount of synthesis of sgRNA
[0048] The obtained sgRNA template was amplified in vitro through the kit to obtain sgRNA in a single-stranded state【 T7 Kit (Ambion)]. The reaction system is as follows:
[0049]
[0050] After mixing, react at 37°C for 12 hours and then purify to obtain sgRNA.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com