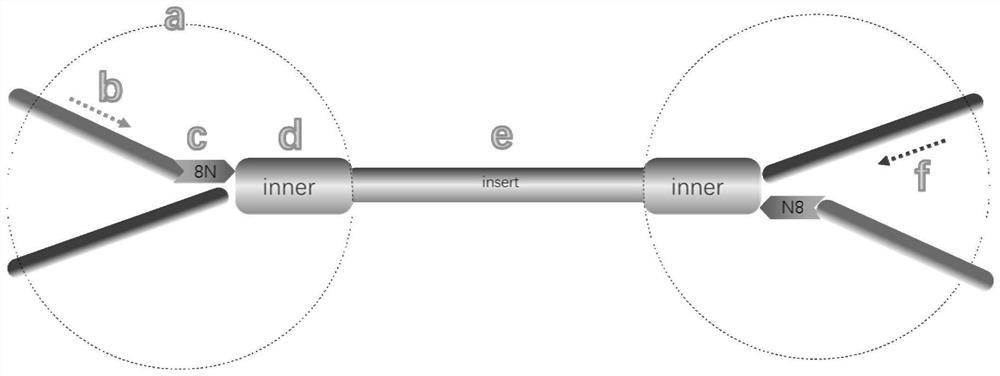
A method for detecting mutation and methylation of tumor-specific genes in ctDNA
A technology of methylation and DNA molecules, applied in the field of biomedicine, can solve the problems of extended detection cycle, low sensitivity of low-frequency mutation detection, low ctDNA mutation content, etc., and achieve the effect of low sample size requirement and great application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0119] Embodiment 1, the construction of MC library
[0120] 1. Methylation-sensitive restriction endonuclease digestion
[0121] Take 5-40ng cfDNA, configure the reaction system as shown in Table 1, and then carry out enzyme digestion treatment in a PCR instrument according to the procedures in Table 2 to obtain enzyme digestion products (stored at 4°C).
[0122] Both Restriction Enzyme and Restriction Enzyme 10×Buffer are products of ThermoFisher. Restriction Enzyme and Restriction Enzyme 10×Buffer can be selected according to different target regions to be tested, and the selection criterion is that the region to be tested contains at least one cleavage site of the methylation-sensitive restriction endonuclease.
[0123] Table 1. Reaction system
[0124] Element volume cfDNA 16.8μl Restriction Enzyme 10×Buffer 2μl Acetylated BSA (10μg / μl concentration) 0.2μl Restriction Enzyme (10U / μl concentration) 1μl total capacity 20μl ...
Embodiment 2
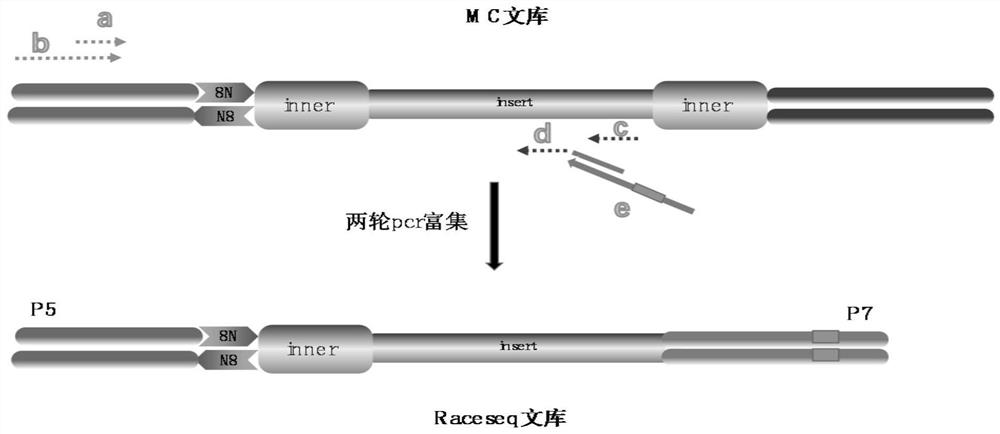
[0165] Example 2, RaceSeq enriches the target region and constructs a sequencing library
[0166] Such as figure 2 As shown, using primers designed for the relevant regions of Chinese hepatocellular carcinoma highly mutated genes (TP53, CTNNB1, AXIN1, TERT), HBV integration hotspot regions and specific hypermethylated regions of hepatocellular carcinoma (EMX1, LRRC4, BDH1, etc.), Cooperate with the fixed primers to perform two rounds of PCR amplification on the MC library, and the amplified product is the sequencing library.
[0167] figure 2 Among them, a is the upstream primer of the first round of library amplification, b is the upstream primer of the second round of library amplification, c is the downstream primer library of the first round of library amplification, which is used for the enrichment of specific target sequences, and d is the second round of library amplification Amplify the downstream primer library for the enrichment of specific target sequences, e is...
Embodiment 3
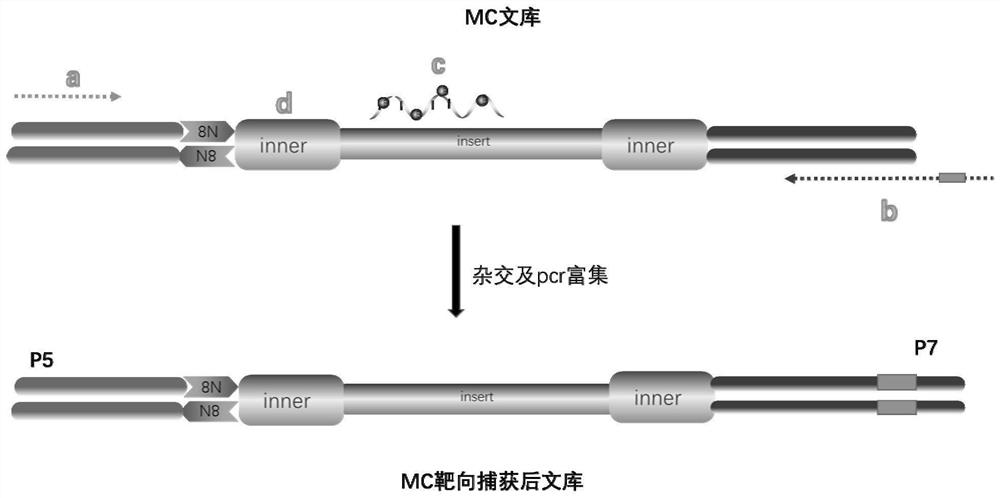
[0212] Example 3, capture and sequencing of MC library
[0213] Such as image 3 As shown, the enrichment of the target region can be captured based on the optimized design of the existing commercial target capture kit. For example: for capture based on methylated regions, please refer to Roche SeqCap Epi CpGiant Enrichment Kit (Roche07138881001) or Illumina Infinium Methylation EPIC BeadChipWG-317-1001), and the design of targeted capture of methylated regions needs to be carried out according to the degree of coverage of restriction sites Screen and adjust probes for bases converted based on bisulfite treatment. The capture based on the gene variation region can refer to the Agilent sureselect XT target capture kit (Agilent5190-8646), and only the primers in the last step of PCR amplification are replaced with the following primers:
[0214] The upstream primer is: 5'-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCT ACACGACGCTCTTCCG AT CT-3' (sequence 403) ( image 3 In "a"), t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com