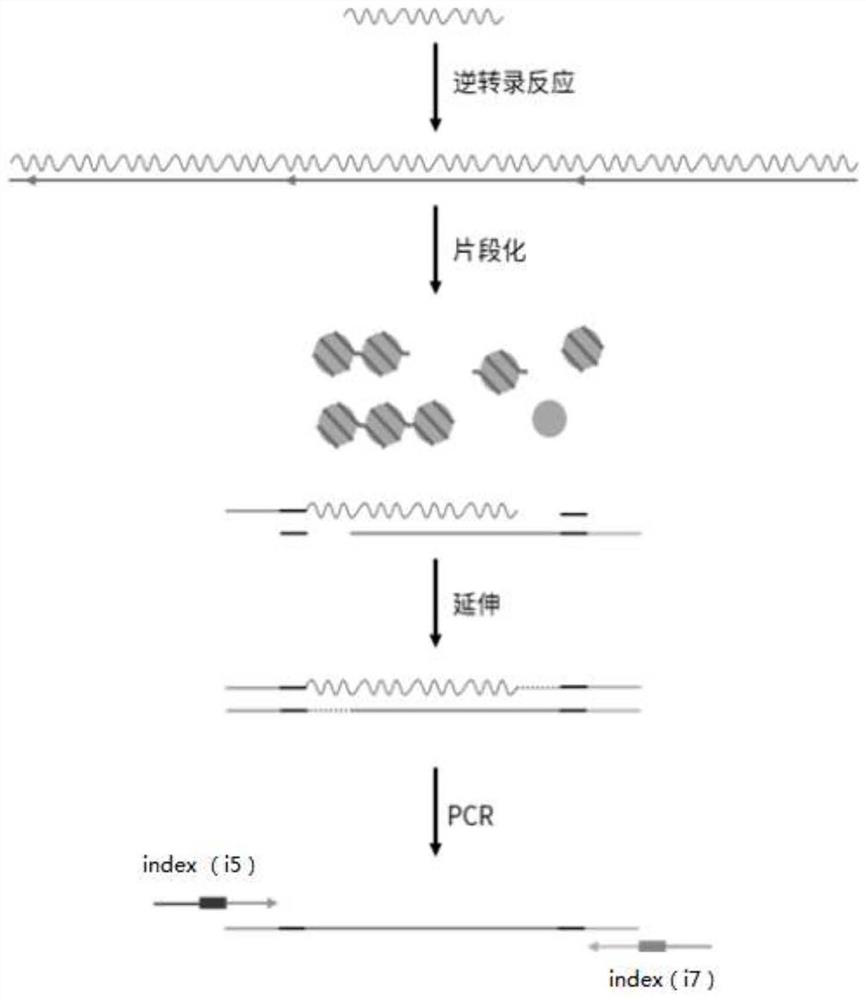
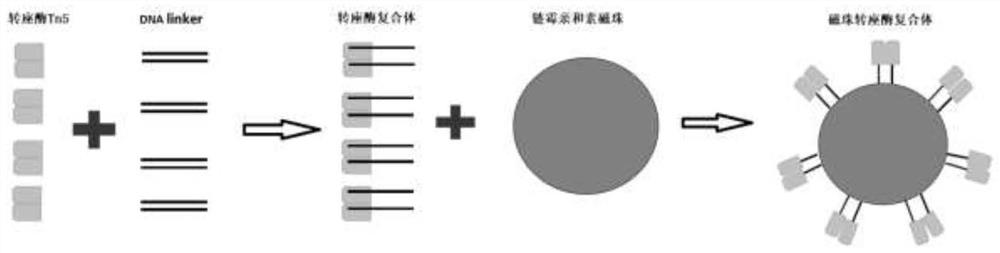
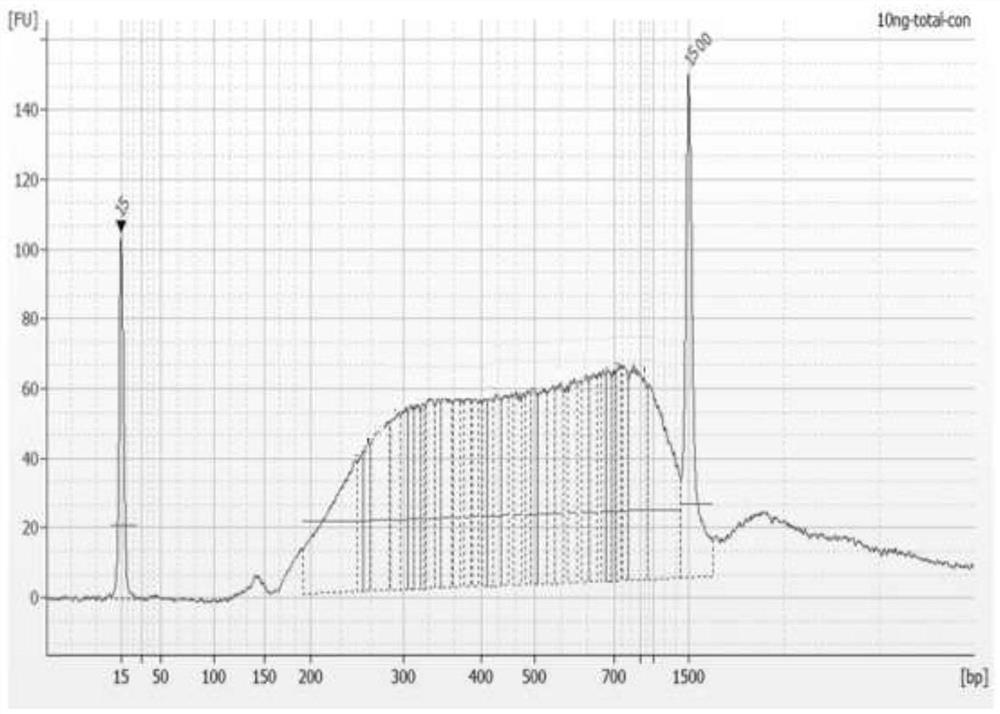
RNA library building method
A library and magnetic bead technology, applied in the field of RNA library construction, can solve the problems of small dosage, complex steps, long library construction period, etc., and achieve the effects of simple experimental steps, flexible input, and reduced dependence.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment example 1
[0036] Implementation Case 1: Assembly of Transposome Complex and Magnetic Beads
[0037] Step 1: DNA linker primer preparation
[0038] The DNA linker primer contains mosaic end sequences recognized by transposase and a sequence connected to magnetic beads. The specific sequence is as follows:
[0039] Biotin-Tn5 ME-A: 5'-NNNNNNNNNN(5-100bp)-TCGTC GGCAG CGTCA GATGT GTATAAGAGA CAG-3' 5'-Biotin
[0040] Biotin-Tn5 ME-B: 5'-NNNNNNNNNNNN(5-100bp)-GTCTC GTGGG CTCGG AGATG TGTATAAGAG ACAG-3' 5'-Biotin Tn5 MErev: 5'-CTGTCTCTTATACACATCT-3' 5'-phosphorylation modification
[0041] Step 2: Assembly of the transposase complex
[0042]The DNA linker annealing buffer, transposase assembly buffer, and Tn5 transposase were provided by the One-step DNA Lib prep kit produced by Wuhan Aibtech Biotechnology Co., Ltd.
[0043] 2.1 DNA linker assembly
[0044] Dissolve the DNA linker sequence in 1xTE buffer, adjust the concentration to 100uM, and assemble the DNA linker with Biotin-Tn5ME-A seq...
Embodiment 2
[0053] Example 2 Building a library with different total RNA inputs in the same species
[0054] 1. Using Oligo(dT) 23 VN for first cDNA synthesis
[0055] Start with total RNA input, respectively input 10ng, 100ng, 1ug mouse total RNA to directly synthesize first cDNA.
[0056] 1. first cDNA synthesis
[0057] 1.1 Take out the Total RNA sample, thaw it on ice, dilute 10ng~1ug sample with Nuclease-free Water to 8μL, and keep it on ice for later use;
[0058] 1.2 Take out 5X First Strand Buffer, Oligo(dT) 23 VN was melted on ice, and the reaction system was prepared according to the following table on ice:
[0059]
[0060]
[0061] *First Strand Synthesis Enzyme Mix components include reverse transcriptase and RNA inhibitor, the reverse transcriptase is ABSScript III reverse transcriptase (ABclonal RK20408) or ABSScript II reverse transcriptase (ABclonalRK21400) or other commercially available reverse transcriptases capable of synthesizing full-length first cDNA r...
Embodiment example 3
[0148] Implementation case 3. Total RNA library construction of different species with the same input amount
[0149] 1. Using Oligo(dT) 23 VN for first cDNA synthesis
[0150] Start with total RNA input, and input the same amount of human, rat, and mouse total RNA respectively to directly synthesize first cDNA.
[0151] 1. first cDNA synthesis
[0152] 1.1 Take out the Total RNA sample, thaw it on ice, dilute 10ng~1ug sample with Nuclease-free Water to 8μL, and keep it on ice for later use.
[0153] 1.2 Take out 5X First Strand Buffer, Oligo(dT) 23 VN was melted on ice, and the reaction system was prepared according to the following table on ice:
[0154] Reagent volume Total RNA sample 8μL 5X First Strand Buffer 4μL Betaine 4μL Oligo(dT)23VN 2μL First Strand Synthesis Enzyme Mix 2μL total capacity 20 μL
[0155] 1.3 Use a pipette to mix evenly, centrifuge briefly, and place the system in a PCR instrument (75°C ho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com