Flammulina velutipes suitable for industrial cultivation and molecular identification method of flammulina velutipes
A technology for classification and naming of Flammulina velutipes, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and measurement/inspection of microorganisms, can solve problems such as irregular fruiting, yellowish color, and low yield per unit area of Flammulina velutipes species, and achieve mushroom production. The effect of white body and small mushroom cap
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
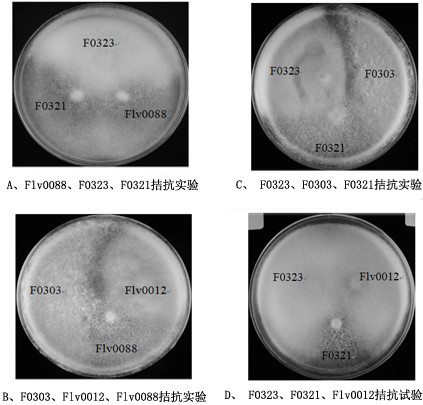
[0016] Embodiment 1: select the Flammulina velutipes 'F0321' and 'F0323' that are mainly planted as hybrid parents, select this test to prepare 180 combinations in total, and have 59 hybrids that contain lock joints through microscopic examination, and screen out 50 of them. After the fruiting test of the better hybrid strains, 11 hybrids capable of fruiting were selected for secondary screening. Among the 11 re-screened hybrid strains, 4 hybrid strains have an average yield of more than 400g, among which Flv0088 has better comprehensive properties and has reached the expected breeding goal. This hybrid strain is named 'Nongwanjin No. 9' and will be used for subsequent batches test.
[0017] Choose Flammulina velutipes 'F0321' and 'F0323' as the hybrid parents, and select 20 single-spore strains of 'F0321' and 20 single-spore strains of 'F0323', which grow faster and have more developed aerial hyphae , paired according to the full combination method, a total of 60 combination...
Embodiment 2
[0024]Example 2: Using SV detection technology to compare the DNA differences between this strain and its parent.
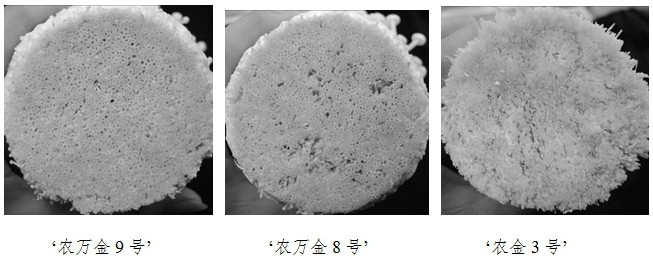
[0025] Test materials: 5 strains of Flammulina velutipes, among which the strain to be tested is 'Nongwanjin No. 9', control variety 1: 'Nongwanjin No. 8' (Flv0012), control variety 2: 'Nongjin No. 3' (F0303), F0321, F0323.
[0026] Inspection method: According to the genome sequencing data, Kmer subtractive assembly technology is used to obtain differential fragments (SV), and several SV sequences are selected accordingly to design specific primers to amplify the samples to be tested. (For references see Alkan, C., Coe, B. P. & Eichler, E. E. Genome structural variation discovery and genotyping. Nature reviews. Genetics, 2011(12), 363–376; Zhu Fengli et al., Whole Genome Resequencing Analysis of Korean Chizhi Zhizhi, World Science and Technology-Modernization of Traditional Chinese Medicine, 21(4)764-774.). SV test primer sequences were synthesized by Qingke...
Embodiment 3
[0056] Example 3: 'Nongwanjin No. 9' has carried out many stable yield tests. The results show that 'Nongwanjin No. 9' meets the requirements of industrial cultivation, and its yield is significantly higher than that of the control variety 'Nongwanjin No. 8' Henongjin No. 3 (F0303).
[0057] Multi-batch stable yield and productive test strains: 'Nongwanjin No. 8', 'Nongjin No. 3' (Flv0303).
[0058] Test location: Fujian Wanchen Biotechnology Co., Ltd.
[0059] Test arrangement: uniform production of strains, factory cultivation at the test site according to local production management methods, bottle cultivation, and multi-zone mobile bed frame cultivation (plastic bottle, 0.43kg (dry material) / bottle). See Table 2 for specific cultivation information:
[0060] Table 2 Multi-batch test information table
[0061]
[0062] Mycelial growth test results: During the experimental seed production period, the mycelium of 'Nongwanjin No. 9' was thicker in PDA, and the optimum gr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com