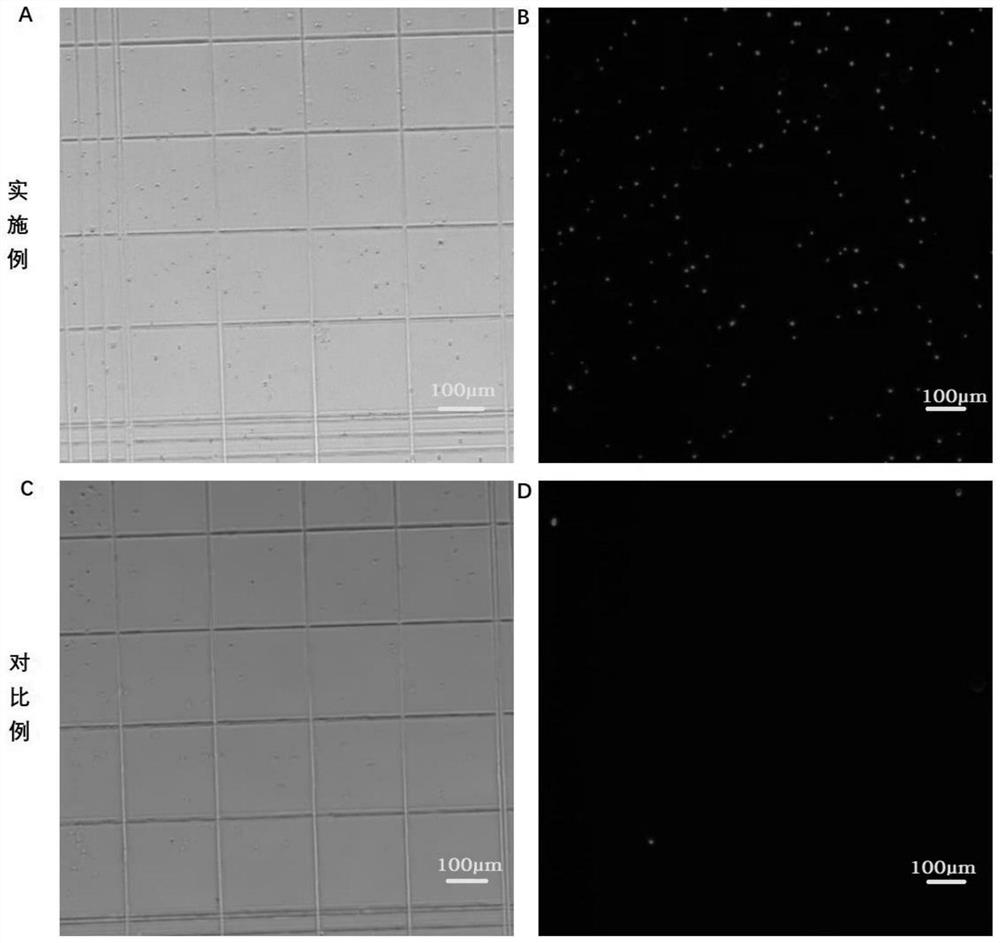
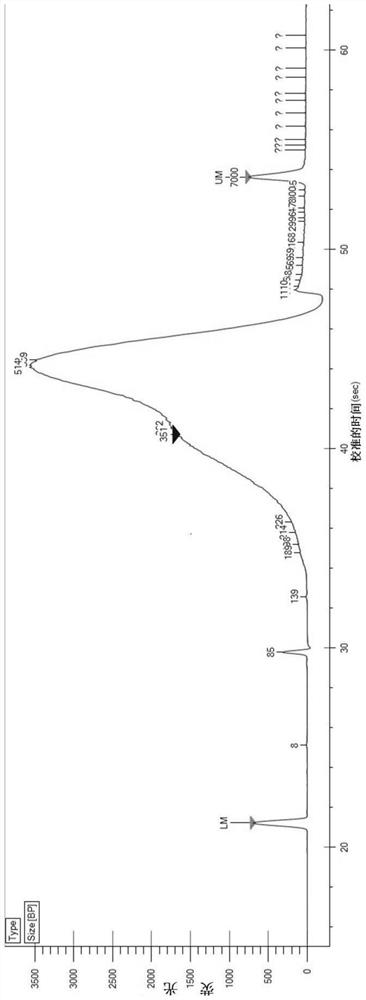
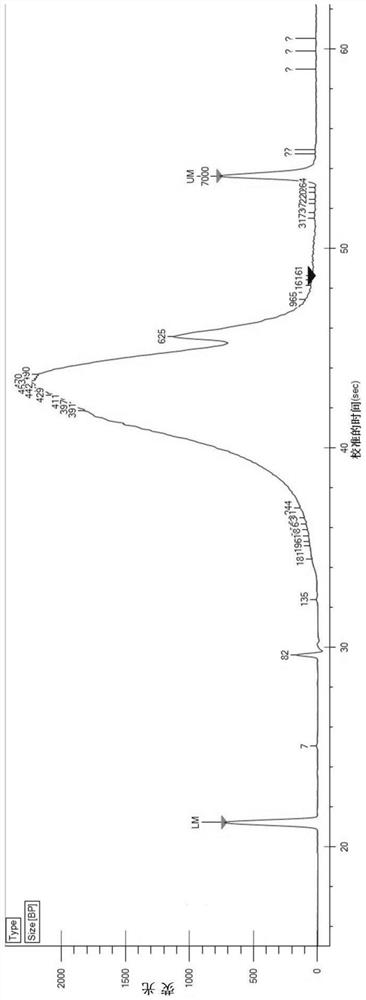
In-vitro tissue cell nucleus separation method for reducing unicellular amplification bias
A cell nucleus and in vitro technology, applied in biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of no quality control result display and instructions, and achieve the effect of saving experimental time, maintaining cell activity, and high simplicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090] (1) Reagent preparation
[0091] 1.WB: 10-20mM Tris-HCl (pH 7.5-7.8), 10-20mM NaCl, 50-100mM KCl, 2-10mM MgCl 2 , 5-15mM CaCl 2 , 0.04% BSA, 1mg / mL PI and 0.2U / μL RNase inhibitor;
[0092] 2.LB: add 2uL NP-40 to 1mL of WB;
[0093] 3. D-Hanks buffer containing 1% FBS, 1 mM DTT.
[0094] (2) Tissue cell lysis
[0095] 1. Pre-cool PBS and prepared WB, LB and NB;
[0096] 2. Pre-cool the plastic petri dish, centrifuge tube, etc. on ice, add 4mL pre-cooled PBS to the petri dish to clean the tissue, and the tissue block should not exceed 3mg;
[0097] 3. Put ~3mg tissue into a pre-cooled centrifuge tube, grind it on ice with a pre-cooled grinding pestle to 0.2mm3 pieces, and place it on ice for 5-10 minutes;
[0098] 4. Blow and suck 10 to 15 times to break up the tissue, and filter it into the flow tube with a 70 μm cell filter;
[0099] (3) Nucleus cleaning
[0100] 1. Centrifuge the nuclear suspension at 500g at 4°C for 5min;
[0101] 2. Remove the supernatant ca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com