Application of the transcription factor hinge1 in the regulation of nitrogen-phosphorus homeostasis in plants
A technology for phosphorus absorption and nucleic acid molecules, applied in angiosperm/flowering plants, applications, plant peptides, etc., can solve problems such as incomplete understanding of signaling pathways
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 Analysis and cloning of HINGE1 gene
[0079] 1. Analysis of HINGE1 gene
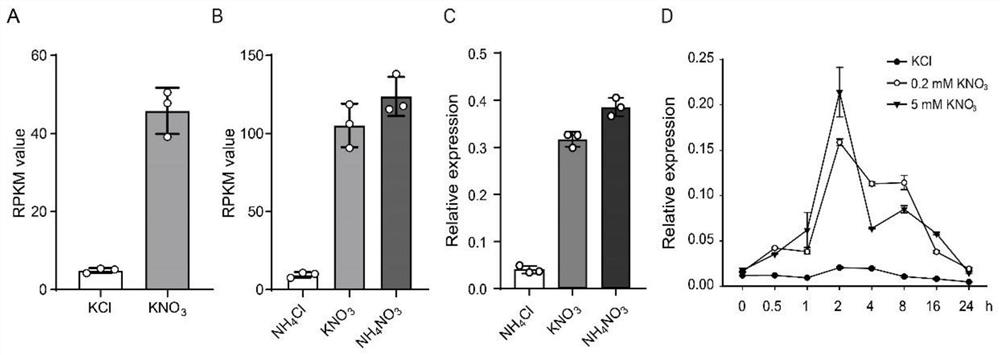
[0080] The inventors first performed transcriptome analysis (RNA-seq) on the seedlings of rice variety ZH11 supplied with different nitrogen sources, and the results are shown in figure 1 B (pure ammonium treatment labeled NH 4 Cl, pure nitrate treatment is marked as KNO 3 , ammonium nitrate 1:1 (calculated as nitrogen, mass ratio) mixed treatment marked as NH 4 NO 3 ); The inventors performed transcriptome analysis (RNA-seq) on rice seedlings induced by short-term nitrate, and the results are shown in figure 1 A (short-term nitrate-inducing treatment labeled KNO 3 , KCl as the control).
[0081] According to the above results, by screening nitrate-induced transcription factors, the inventors obtained a gene (LOC_Os04g56990) that was induced the strongest by nitrate in roots, which encoded the GARP class MYB family transcription factors, and named it HINGE1 (Highly Induced by Nitr...
Embodiment 2
[0110] Embodiment 2, expression analysis of HINGE1 gene
[0111] 1. Expression analysis of rice HINGE1 supplied with different nitrogen sources
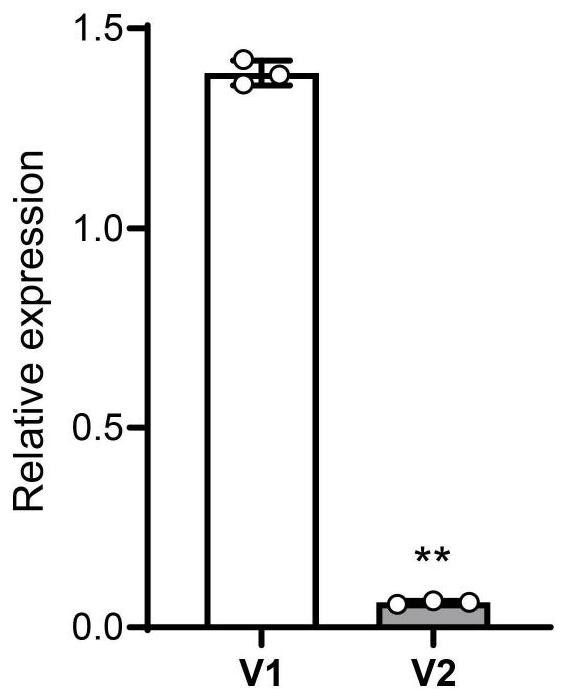
[0112] The inventors used qRT-PCR (primers are qHINGE1-1F and qHINGE1-1R) to analyze the expression of HINGE1 in rice ZH11 seedlings supplied with different nitrogen sources, and the results are shown in figure 1 At the same time, the expression of HINGE1 in rice seedlings induced by different concentrations (0mM, 0.2mM, 5mM) of nitrate in a short period (0.5, 1, 2, 4, 8, 16, 24 hours) was analyzed, and the results are shown in figure 1 D. The above results all proved that the expression of HINGE1 was induced by nitrate.
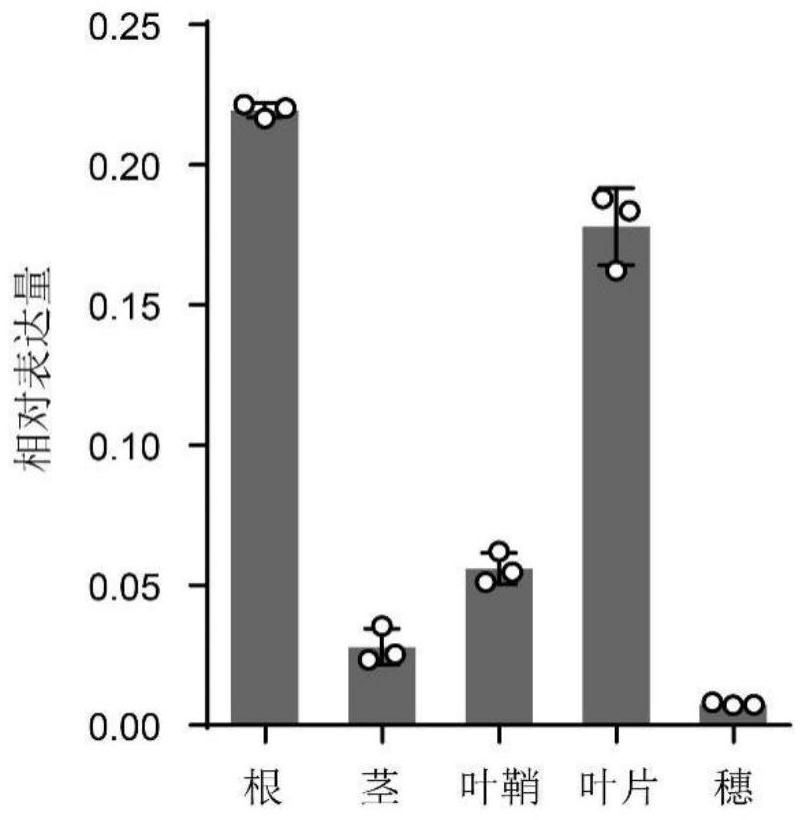
[0113] 2. Analysis of HINGE1 expression in different rice tissues
[0114] 2.1qRT-PCR analysis
[0115] The expression of HINGE1 in different tissues of rice was further analyzed by qRT-PCR, the primers used were qHINGE1-1F and qHINGE1-1R, and it was found that the expression of HINGE1 was the highest in roots ...
Embodiment 3
[0127] Example 3, HINGE1 transcription factor characteristic analysis
[0128] 1. Subcellular localization analysis
[0129] Transcription factors generally perform their transcriptional activation or repression functions in the nucleus, and the inventors analyzed the subcellular localization of HINGE1. Therefore, the inventor constructed a plasmid in which eGFP (enchanced GFP) was fused to the C-terminus of the HINGE1 protein driven by the 35S promoter, and transformed rice protoplasts. The specific steps were as follows:
[0130] 1.1 Construction of HINGE1-eGFP vector
[0131] Primers containing In-Fusion adapters were designed for the pCAMBIA2300-35S-eGFP(C) vector, and the primer sequences are as follows:
[0132] HINGE1-GFP-F:5' (The sequence indicated by the underline is the KpnI enzyme recognition site sequence, and the sequence indicated by the bold font is the In-Fusion linker sequence);
[0133] HINGE1-GFP-R:5' (The sequence indicated by the double underline i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com