OsFLP protein related to plant salt tolerance as well as related biological materials and application thereof
A biomaterial and protein technology, applied in angiosperms/flowering plants, applications, plant peptides, etc., can solve problems such as the intricate regulation network of plant adversity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1. Acquisition of plant salt-tolerance-related gene OsFLP
[0074] 1. Sequence analysis of rice OsFLP protein
[0075]Using the amino acid sequence of Arabidopsis AtFLP in NCBI (http: / / www.ncbi.nlm.nih.gov / ) and the rice genome database (http: / / rice.plantbiology.msu.edu / ), to find Homologous proteins of Arabidopsis AtFLP. As a result, a protein having 33.89% similarity to AtFLP's amino acid sequence was obtained and named as OsFLP, and its amino acid sequence is shown in Sequence 1 in the Sequence Listing.
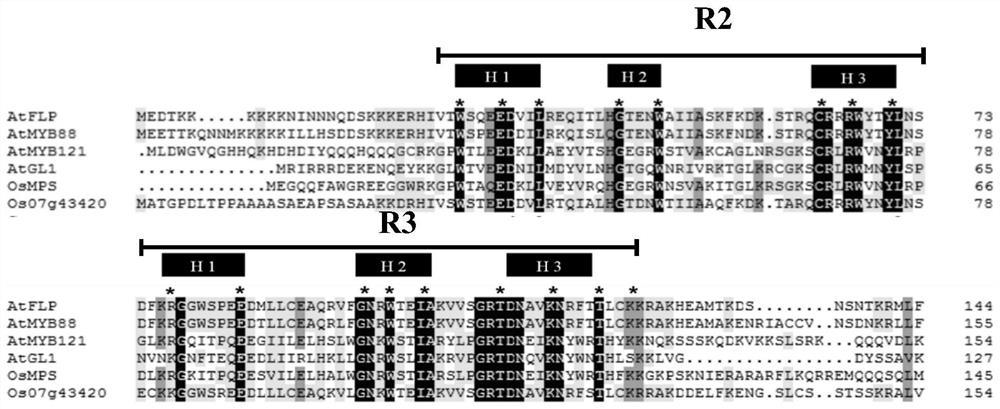
[0076] Through comparative analysis with several known R2R3-MYB family protein sequences, including Arabidopsis AtFLP, AtMYB88, AtMYB121, ATMYB0 (ATGL1) and rice OsMPS protein sequences, the results are shown in figure 1 , found that there are two MYB domains, R2 and R3, at the N-terminal of OsFLP protein. Among them, R2 contains three conserved motifs of H1, H2 and H3, and R3 contains three conserved motifs of H1, H2 and H3, which proves that OsFLP is a ...
Embodiment 2
[0093] Embodiment 2, the acquisition of gene OsFLP mutant
[0094] 1. Using TILLING technology to obtain osflp-1 mutants
[0095] TILLING (Targeting Induced Local Lesions IN Genomes) technology is a new reverse genetics research method, using the chemical mutagen ethyl methanesulfonate to mutate the seeds, and then using high-throughput Sequencing methods quickly and efficiently identify point mutations from mutagenized populations. In order to better analyze the function of OsFLP in rice, we used TILLING technology to obtain osflp mutant lines at different mutation sites, see Table 1 for details. We observed and compared the obtained strains at different mutation sites, and found a strain A2349-2 with a different stomatal phenotype from the wild type, and named the mutant osflp-1 mutant.
[0096] Table 1
[0097]
[0098]
[0099] 2. Mutation site analysis of osflp-1 mutant
[0100] We designed corresponding primers to amplify OsFLP genomic DNA in ZH11 and osflp-1 m...
Embodiment 3
[0101] Embodiment 3, the acquisition of transgenic OsFLP rice
[0102] 1. Construction of OsFLP gene overexpression vector and acquisition of rice transgenic materials
[0103] OsFLP gene overexpression vector is using pH7WG2D.1 (plasmid map see Figure 5 ) as a starting carrier, adopting the gateway system method to construct, the specific implementation is as follows: amplify the cDNA fragment of OsFLP (the nucleotide sequence is sequence 2), after gel recovery, connect the cDNA fragment of OsFLP to the entry vector TOPOvector, and name the obtained plasmid is OsFLP-TOPO. Digest the plasmid OsFLP-TOPO with PVUⅠ restriction endonuclease (recognition site: CGATCG, the cut fragment is sticky, it belongs to restriction endonuclease type II) at 37°C for 3 hours, and put The enzyme products were separated by agarose gel electrophoresis and recovered by cutting the gel. Mix 1.5 μL of recovered product, 0.5 μL of pH7WG2D.1 plasmid, and 0.5 μL of Gateway LR ClonaseⅡenzyme mix, and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com