Nucleotide sequence for encoding novel coronavirus antigen and application of nucleotide sequence
A nucleotide sequence, coronavirus technology, applied in the field of vaccines
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: Optimization of S protein-encoding nucleic acid and vector construction
[0044] 1. The principle of nucleic acid sequence optimization is: (1) optimize the degenerate codons according to the host cell’s preference for nucleic acid codons, so that the optimized sequence contains more nucleic acid codons that are conducive to host cell recognition; On the basis of codon preference optimization, further optimize the GC content in the nucleic acid sequence, so that the GC content-optimized sequence can express more target proteins; (3) optimize the nucleic acid sequence so that it can transcribe more stable mRNA, which is beneficial to the target protein. (4) Change the codon frequency of host preference and increase the CAI index (codon adaptation index).
[0045] Optimization purpose: to increase the protein expression of the target protein in host cells.
[0046] Optimization strategy: optimize the third base of the amino acid coding codon from A to C or G, ...
Embodiment 2
[0051] Embodiment 2: Construction of nucleic acid vaccine (sequence after each optimization)
[0052] 1. Nucleic acid vaccine candidate sequence transformation
[0053] (1) Take 100 μL of competent cell (such as DH5α) suspension from a -70°C refrigerator and thaw on ice.
[0054] (2) Add the plasmid DNA solutions prepared in Example 1 (volume no more than 10 μL): pVAX1-S (WT), pVAX1-ADV400, pVAX1-ADV401 and pVAX1-ADV402 into competent cells, shake gently , placed on ice for 30 minutes.
[0055] (3) Heat shock in a water bath at 42°C for 90 seconds, then quickly place on ice to cool for 5 minutes.
[0056] (4) Add 1 mL of LB liquid culture medium (without antibiotics) to the tube, mix well and incubate with shaking at 37°C for 45 minutes to restore the bacteria to normal growth state.
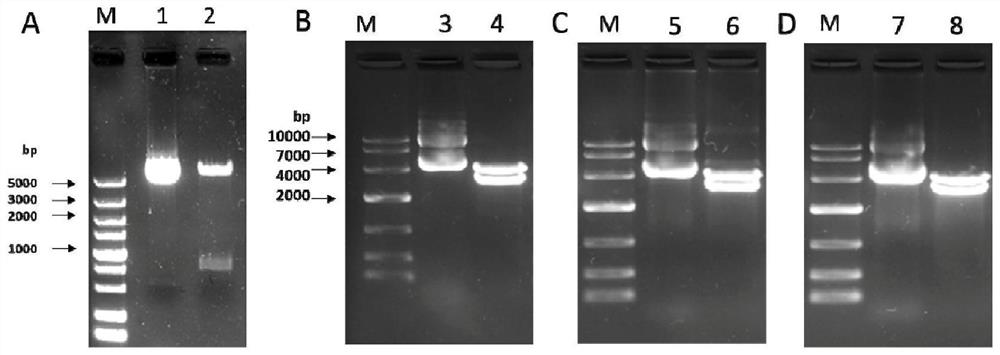
[0057] (5) Shake the above-mentioned bacterial liquid and apply 100 μL on a screening plate containing appropriate antibiotics, place it face up, and invert the culture dish after the bacteri...
Embodiment 3
[0072] Example 3: Identification of Candidate Nucleic Acid Vaccine Mammalian Cell Transcription
[0073] 1. In vitro transfection of nucleic acid vaccine
[0074] (1) Take out the frozen HEK293 cell line (ATCC, CRL-3216) from liquid nitrogen, centrifuge at 1000rpm for 5min after 37℃ water bath to remove DMSO;
[0075] (2) Add serum-free DMEM culture solution to wash once, in 5 mL of DMEM culture solution containing 10% calf serum, 37°C, 5% CO 2 nourish. 24 hours before transfection, cells were digested with trypsin (0.25%) at 37°C for 5 minutes and terminated, with (1-3)×10 5 The density of cells / well was spread on 6-well plate or 35mm culture dish, add 2mL growth medium and 37℃, 5%CO 2 Cultivate in the incubator for 20-24 hours until the cell density is 50-80%;
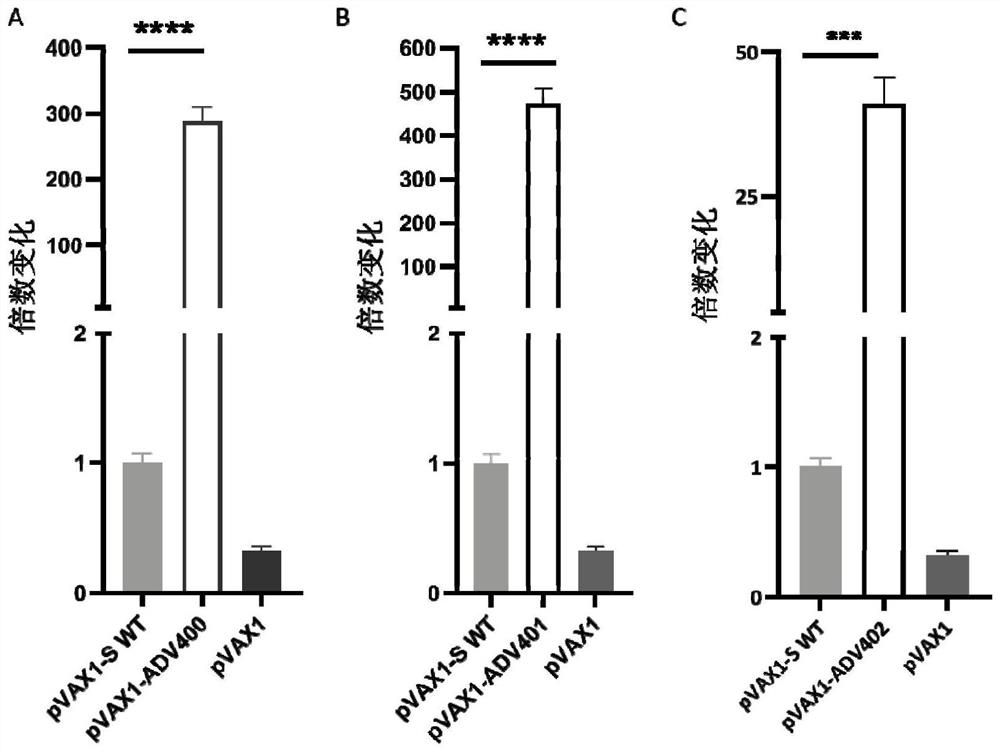
[0076] (3) Simultaneously transform pVAX1-ADV400, pVAX1-ADV401 and pVAX1-ADV40, wild-type pVAX1-S (WT), and empty plasmid pVAX1 for parallel experiments. Specifically, take 2 μg of the aforementioned 5 plasmids, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com