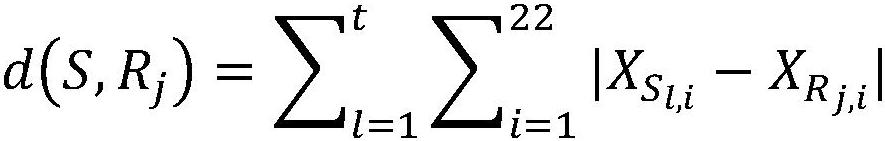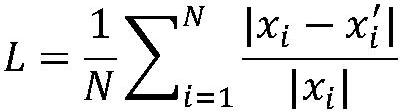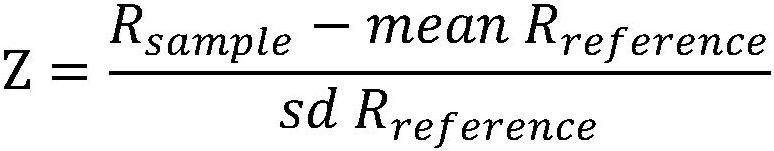Non-invasive prenatal detection device
A sample and data correction technology, applied in the field of bioinformatics, can solve the problems of trivial data segmentation and non-provision, and achieve the effects of reducing detection costs, avoiding false negatives or false positives, and shortening the detection cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] This embodiment provides a method for detecting chromosomal variation using the non-invasive prenatal detection device of the present invention.
[0051] The device includes: a detection module, a data quality control and preprocessing module, a data correction and processing module, and a judging module. Specific steps are as follows:
[0052] 1. Negative reference set construction
[0053] (1) Sample selection
[0054] A total of 9,000 pregnant women whose gestational age was greater than or equal to 12 weeks and had no chromosomal abnormalities in karyotype analysis were selected, and there was no significant difference in the proportion of males and females. Cell-free DNA was extracted, genome sequencing was performed according to high-throughput methods, and fastq data with a read length of 50 bp was obtained by single-end sequencing. The sequencing platform used was the MGISEQ-2000 gene sequencer manufactured by MGI.
[0055] (2) Data preprocessing
[0056] Th...
Embodiment 2
[0100] In this embodiment, the method shown in Example 1 and the comparative method are used to test the sample to be tested.
[0101] (1) The peripheral blood of 20 pregnant women was selected for testing, numbered S1-S20, and the karyotype results showed that one pregnant woman had a fetus with trisomy 21, and the rest of the samples were negative.
[0102] (2) The 20 samples were tested according to the method of Example 1 of the present invention and the comparative method. The comparison method is the same as the method in Example 1, the only difference is that the construction and use of the dynamic database is not carried out, and the deviation caused by different chromosome baselines is not re-corrected, and all negative reference sets are used. In the comparison method, when the number of samples with |Z| greater than or equal to 3 in the same batch is more than half of the total sample data in this batch, and the Z values of the same chromosome are all too large or...
Embodiment 3
[0113] In this embodiment, the method shown in Embodiment 1 is used to test the sample to be tested.
[0114] (1) The peripheral blood of 30 pregnant women was selected for testing, numbered T1-T30, and the karyotype results showed: 19 cases of trisomy 21, 3 cases of trisomy 18, and 1 case of trisomy 13 Body syndrome, 7 cases of microdeletion and microduplication syndrome.
[0115] (2) Using Example 1 to detect each sample.
[0116] Table 4 shows the detection results of 23 cases of abnormal aneuploidy samples (see the bold data in the table), all of which are consistent with the karyotype results. Table 5 shows the detection results of 7 cases of microdeletion and microduplication syndrome samples, all of which are consistent with the karyotype results, from which it can be seen that the fetal concentration is 0.052, and the region with a fragment size of about 2M can also be accurately detected.
[0117] Table 4 Abnormal results of aneuploidy
[0118] sample numb...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com