UGT enzyme activity detection method and application thereof
A detection method and technology for enzyme activity, applied in the field of medicine, can solve the problems of low detection throughput, cumbersome and complicated operation, and inability to screen and evaluate multiple UGT subtype enzymes on a large scale, achieving good repeatability, convenient operation, and low cost. The effect of the cost of experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1. Broad-spectrum research on the metabolism of Ophiopogon japonicus high isoflavone A by human UGT enzyme
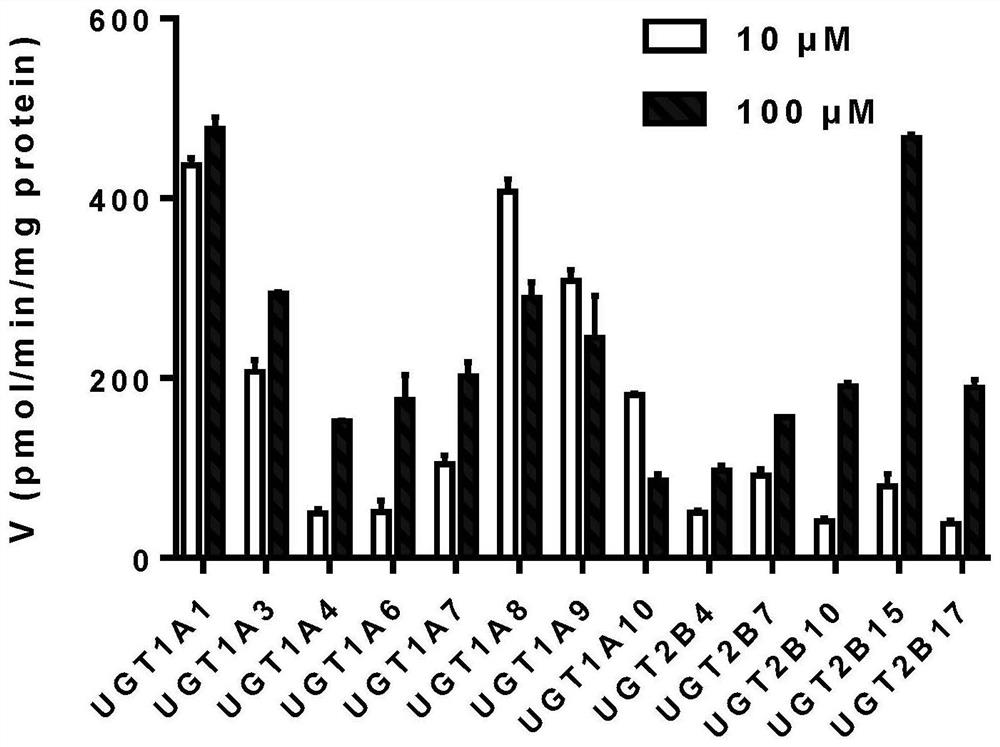
[0038]Phenotype study of UGT enzymes involved in glucose-binding metabolism of broad-spectrum substrate Ophiopogon japonicus high isoflavone A. Using 13 commercialized recombinant human UGT single enzymes (UGT1A1, UGT1A3, UGT1A4, UGT1A6, UGT1A7, UGT1A8, UGT1A9, UGT1A10, UGT2B4, UGT2B7, UGT2B10, UGT2B15, UGT2B17) as the target UGT enzyme source, the substrate MOA Metabolic characterization. details as follows:
[0039] (1) Mix Tris-HCl buffer (50mM, pH=7.4) and magnesium chloride (5mM), and use one of the 13 UGT single enzymes as the target UGT enzyme (0.05mg / ml recombinant human UGT single enzyme), Ophiopogon japonicus high isoflavone A (concentration uniformly 10 μM and 100 μM) was mixed in the EP tube, and pre-incubated at 20-60°C for 3-5 minutes, and the pH value of the incubation system was 5-10. Concrete reaction condition is as follows:
[0040]...
Embodiment 2
[0045] Example 2. Preparation and identification of Ophiopogon japonicus high isoflavone A metabolites by UGT enzymes
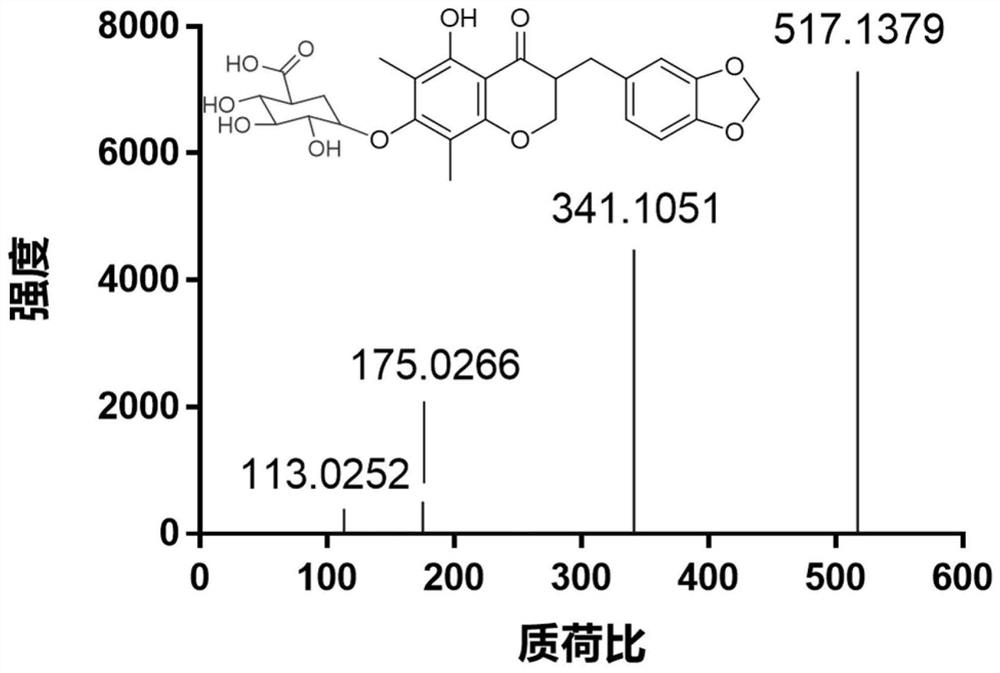
[0046] After using high-resolution mass spectrometry to analyze and identify the metabolites of Ophiopogon japonicus high isoflavone A catalyzed by UGT (such as figure 2 ), a large number of preparations and structural characterizations of its product MOAG were carried out by using biological preparation methods and NMR techniques, respectively. (As shown in Figure 6). Specific steps are as follows:
[0047] (1) Ophiopogon japonicus high isoflavone A (8.52mg), Tris-HCl buffer solution (50mM, pH 7.4), MgCl 2 (5mM), polyoxyethylene hexadecyl ether (0.1mg / mg protein) and 13 recombinantly expressed UGT enzymes (1.85μL for each enzyme, final protein concentration of 0.8mg / ml) were mixed. After 5 min pre-incubation at 37°C, UDPGA (2 mM) was added to start the reaction.
[0048] (2) After incubation at 37°C for 6 hours, the reaction was terminated by adding ice...
Embodiment 3
[0052] Embodiment 3. Stability investigation
[0053] (1) Mix Tris-HCl buffer (50mM, pH=7.4) with MgCl under physiological conditions 2 (5mM), with 13 kinds of UGT enzymes expressed recombinantly (0.31 μL for each enzyme, the final protein concentration is 0.1mg / ml) as the enzyme source, and the substrate Ophiopogon japonicus high isoflavone A solution (concentration selection 1 / 10~1K m ) were mixed in a centrifuge tube, the reaction temperature was 37°C, and the pH of the incubation system was 7.4.
[0054] (2) Pre-incubation: incubate the mixture in (1) at 37° C. for 3-5 minutes to ensure sufficient contact between the enzyme and the substrate Ophiopogon japonicus high isoflavone A.
[0055] (3) Add UDPGA and the reaction time is 30 minutes.
[0056] (4) Samples were taken and analyzed after being placed at 4°C for 0 hour, 24 hours and 48 hours to investigate its stability under the storage condition of 4°C refrigerator. The result is as image 3 shown.
[0057] The res...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com