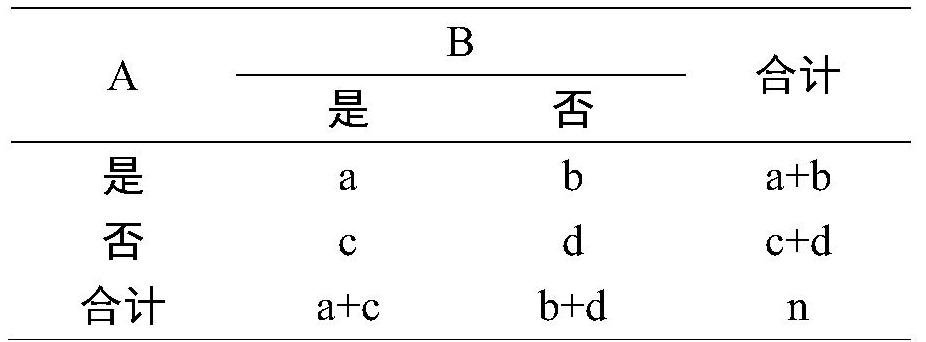
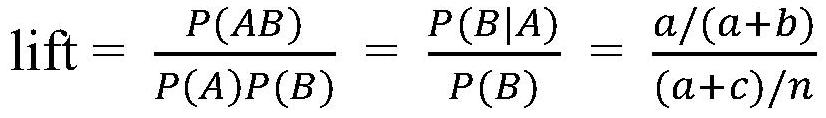Method for screening disease phenotype related mutation sites and application thereof
A technology of mutation sites and phenotypes, applied in the field of bioinformatics, can solve the problems of no observation value of genome combination, sparse data distribution, and dimension confusion, avoiding the influence of allele frequency, high efficiency, and reducing the total sample size. amount of effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] In this embodiment, the method for screening disease phenotype-related mutation sites provided by the present invention is used to mine type 2 diabetes-related SNP sites, as follows:
[0055] 1. Select 200 patients with type 2 diabetes and 200 normal people as controls for microarray sequencing, with a total of 743,722 loci.
[0056] 2. Association rule analysis: According to the genotype of the mutation site, the mutation data and sample phenotype data are converted into binary variables, and the association rule analysis parameters are set. The minimum support degree min_sup=20%, the minimum confidence degree min_conf=80% .
[0057] 3. Apply FP-Growth algorithm to generate frequent itemsets.
[0058] 4. After obtaining the frequent itemsets, find the association rules whose confidence is greater than min_conf as strong association rules
[0059] 5. Select the effective strong association rules from the strong association rules, that is, select all the rules whose ac...
Embodiment 2
[0071] In this example, 100 cases of hypertension, 126 cases of obesity, 410 cases of lung cancer, 360 cases of breast cancer, 134 cases of colorectal cancer and 200 normal samples were selected for GWAS analysis and association rule analysis, and the p-value in the GWAS analysis results was selected <10-e7 and the top 20 sites with p-value<0.005 in the analysis of association rules, compare the proportion of the detected sites in the GWAS Catalog database and record the phenotype-related sites The results are shown in Table 5:
[0072] table 5
[0073]
[0074] It can be seen that the number of SNP sites obtained by analyzing each phenotype using association rules has a higher proportion of the phenotype-related sites recorded in the GWAS Catalog database than the GWAS analysis results.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com