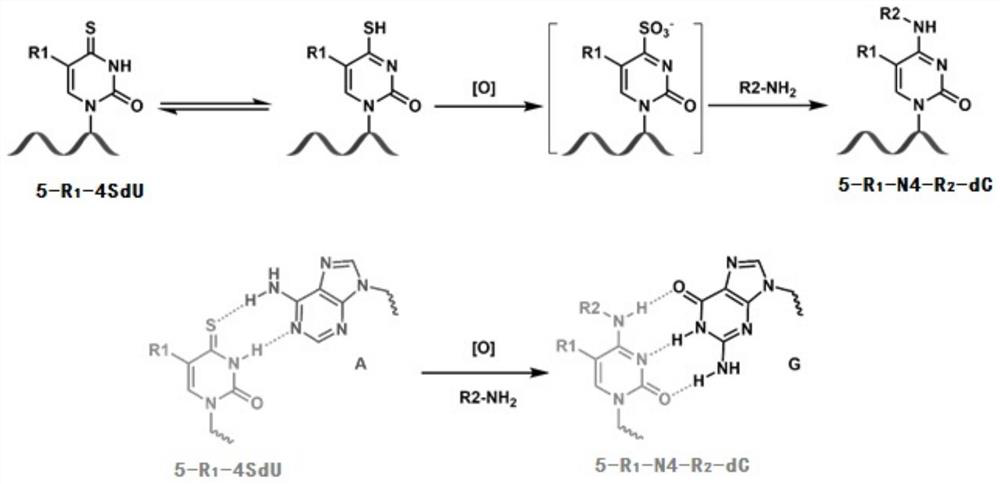
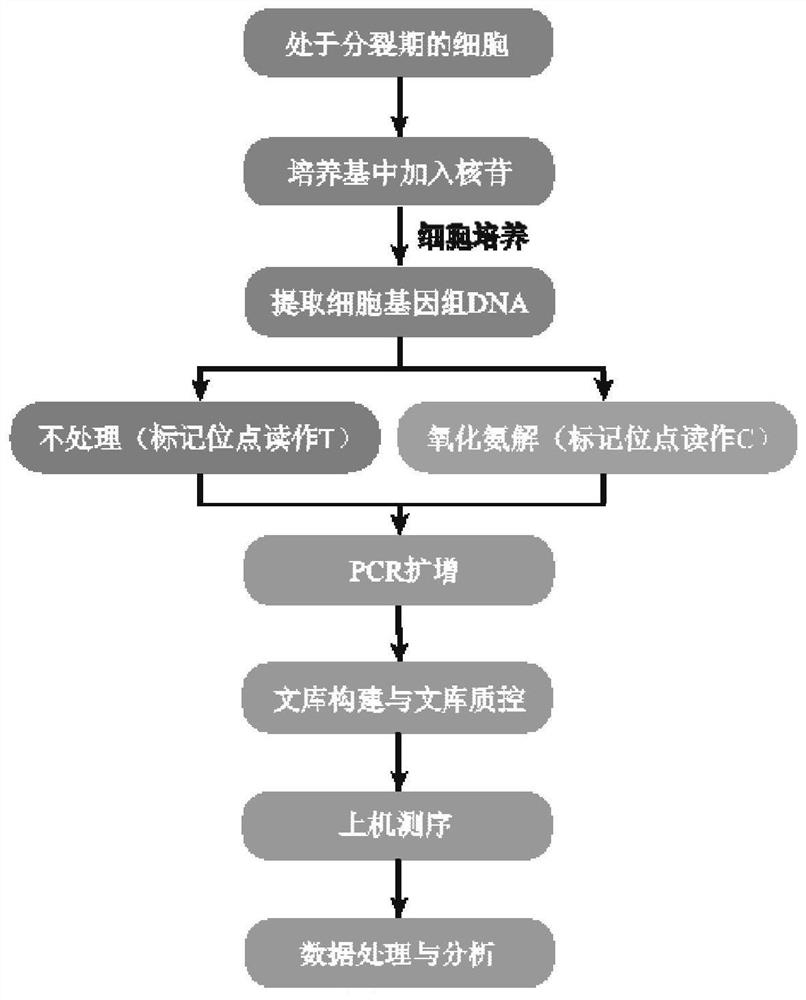
Nucleic acid metabolism marker detection method based on 4-thio-nucleoside oxidative aminolysis reaction and sequencing technology
An oxidation reaction, nucleic acid technology, applied in the field of biochemistry, can solve the problem of low resolution
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 14
[0117] Example 14 ST carries out DNA metabolic labeling to cells
[0118] Use ultrapure water to prepare 4ST stock solution, and filter it with a sterile water phase filter membrane. Store the stock solution at -20°C and preheat it in a constant temperature incubator at 37°C before use. MCF-7 cells in the dividing phase or after injury were cultured in Dulbecco's high-glucose medium containing 10% fetal bovine serum, and 4ST stock solution was added to the medium to make the final concentration of 4ST in the medium 1 mM. Place cells at 37°C, 5% CO 2 Continue culturing in the incubator for a period of time, so that the cellular DNA will incorporate 4ST into it during the replication or repair process. Cell DNA was isolated for subsequent in vitro experiments, the culture medium was discarded, cells were obtained, and cell DNA was extracted using a DNA extraction kit.
Embodiment 2
[0119] Embodiment 2 Oxidative Amination Reaction Treatment of Model Nucleic Acid or Cellular DNA Containing 4ST
[0120] Add 50 pmol of model nucleic acid or 500 ng of cell DNA obtained in Example 1 to a 0.2 mL flat-cap PCR tube, and add EDTA and 100 mM NH at a final concentration of 1 mM in sequence. 4 Ac-NaOH (pH 9.0), and finally add NaIO at a final concentration of 400 μM 4 , the total volume of the reaction system is 40 μL. After adding NaIO 4 Immediately thereafter, place the PCR tube in the PCR instrument, set the temperature program, react at 80°C for 3 hours, and drop the temperature to 4°C at the end of the reaction. After the reaction, the nucleic acid was centrifuged through an ultrafiltration tube to remove and purify small molecules. Wherein, the amount of model nucleic acid added is 100 pmol / 40 μL, and the amount of cell DNA added is 500 ng / 40 μL. NH 4 The concentration of ammonia produced by Ac-NaOH was 100 mM, and the pH of the reaction system was 9.0. N...
Embodiment 3
[0121] Example 3 Two PCRs of Model Nucleic Acids to Prepare Generation Sequencing Samples
[0122] Using untreated 4ST-60nt and 4ST-60nt after oxidative amination reaction as templates, primer1F and primer1R shown in Table 1 as a pair of primers, and Taq DNA polymerase for the first PCR: PCR with 0.2mL flat cap Template DNA with a final concentration of 10 nM, a pair of primers with a final concentration of 0.5 μM, and 25 μL of Taq HS mix were added to the tube to make the volume of the PCR system 50 μL. Place the PCR tube in the PCR instrument, set the PCR program as 94°C for 3min, [94°C for 10s, 52°C for 30s, 72°C for 1min] for 30 cycles, 72°C for 8min, 4°C∞. 5 μL of the PCR product was identified by 1.5% agarose gel electrophoresis, and the remaining solution was purified with E.Z.N.A.Gel Extraction Kit to remove polymerase, dNTP, unreacted primers, etc., and finally obtained 119bp double-stranded DNA.
[0123] Use the purified first PCR product as a template, primer2F and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com