Method for obtaining cucumber germplasm material with high powdery mildew resistance through multi-gene editing
A technology for powdery mildew and cucumber powdery mildew, applied in the fields of biotechnology and molecular biology, can solve the problems of chain burden, bad traits, huge screening workload, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
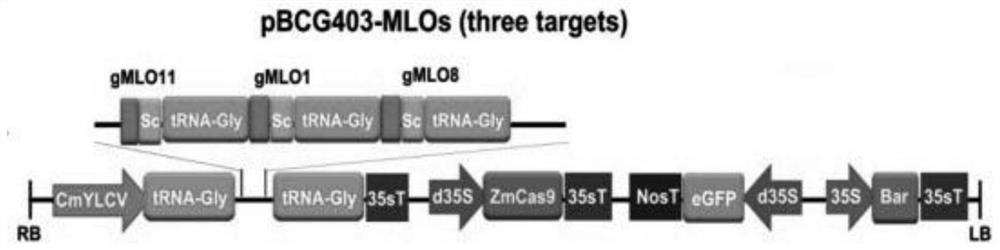
[0050] 1.1 Construction of CRISPR / Cas9 expression vector and Agrobacterium transformation
[0051] 1. Design of sgRNA
[0052] Use Geneious software to select the exon regions of CsMLO1, CsMLO8, and CsMLO11 genes (Cucumber (Chinese Long) v3Genome) for sgRNA design: the sgRNA sequence has a 5'-(N)20-NGG-3' structure (N is A, G Any one of the four bases, C, T), and the GC content is between 40% and 70%.
[0053] The cucumber CsMLO1 sgRNA sequence is shown in SEQ ID NO.4, the CsMLO8 sgRNA sequence is shown in SEQ ID NO.5, and the CsMLO11 gene sgRNA sequence is shown in SEQ ID NO.6.
[0054] 2. Primer Design and PCR Amplification
[0055] (1) Design primer sequences according to the sgRNA of CsMLO1, CsMLO8, and CsMLO11 genes. The primer sequences are as follows:
[0056] Primer name Primer sequence (5'-3') CsMLO11-F SEQ ID NO.7 CsMLO1-R SEQ ID NO.8 CsMLO1-F SEQ ID NO.9 CsMLO8-R SEQ ID NO.10 CsMLO8-F SEQ ID NO.11 VECTOR-R SE...
Embodiment 2
[0079] 2.1 Transformation of Agrobacterium
[0080] The correctly sequenced expression vector plasmid returned by Qingke Company was transformed into Agrobacterium EHA105 to prepare for the subsequent cucumber genetic transformation infection experiment. Take 2 μL of the plasmid in the ultra-clean workbench, add it to 100 μL of EHA105 Agrobacterium competent cells, flick and mix well. After mixing, ice-bath for 5 minutes, freeze in liquid nitrogen for 1 minute, and then quickly place in a 37°C water bath for 5 minutes. Add 800 μL of LB medium, and incubate at 28° C. on a shaker at 220 rpm for 2 hours. Centrifuge at 12,000 rpm for 1 min to concentrate the bacterial solution, add 200 μL LB liquid medium to resuspend, mix well, spread it evenly on LB+Kana solid medium, and culture in dark at 28°C for 2 days. Pick out the monoclonal plaques of Agrobacterium and shake the bacteria with LB+kana+rif liquid for 12 hours, and store the bacteria solution in a -80°C ultra-low temperatu...
Embodiment 3
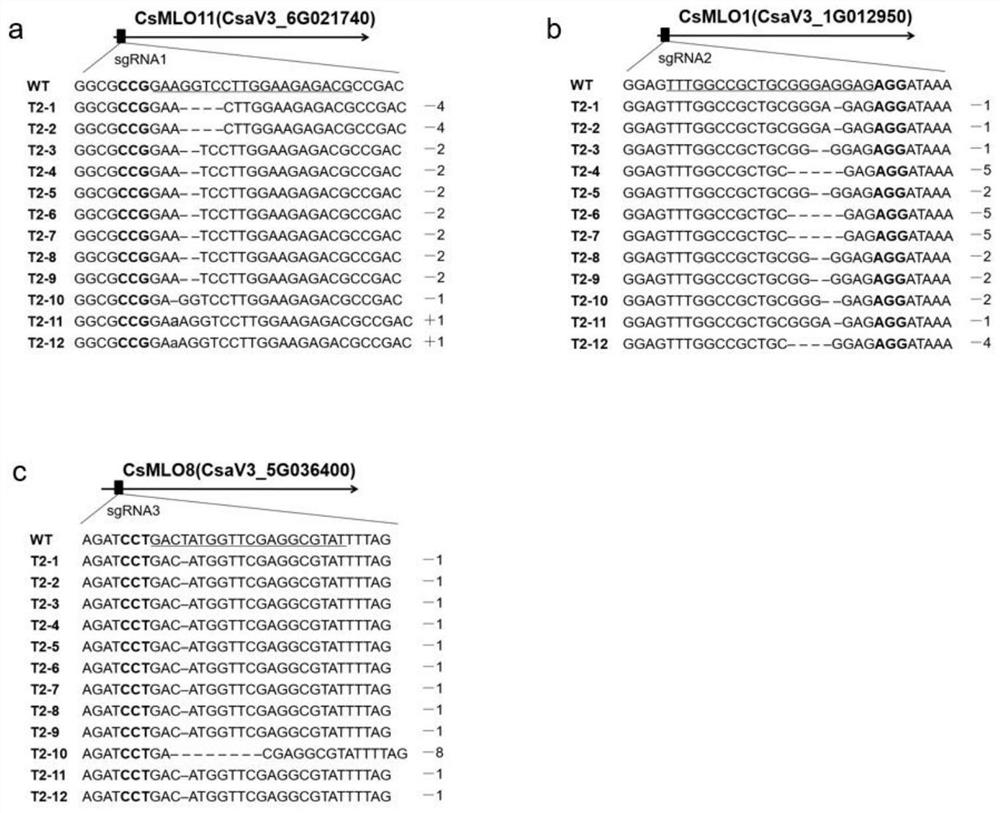
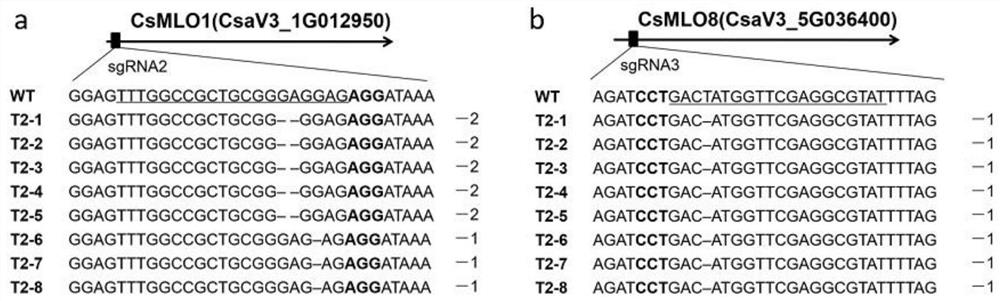
[0090] Example 3, DNA extraction and detection of gene-edited cucumber plants
[0091] 3.1. Extraction of DNA from gene-edited cucumber plants
[0092] The DNA of the T2 generation transgenic cucumber leaves in Example 2 was extracted by using the CTAB method. The method specifically includes the following steps: take a small amount of cucumber leaves about 0.2-0.5g into a 2mL centrifuge tube, add clean steel balls and 1mL of 2% CTAB solution, and grind to powder with a sample mill (50Hz, 30s). The ground sample was placed in a water bath at 65 °C for 1 h. Invert the centrifuge tube several times during this time. Centrifuge at 12000rpm for 10min at 25°C. Pipette the supernatant (approximately 800 µL) into a new 2 mL centrifuge tube. Add an equal volume of chloroform / isoamyl alcohol (24:1) into a fume hood and mix by inverting, and centrifuge at 12000 rpm (4°C) for 15 min. Carefully draw the supernatant (about 600 μL), add it to a 2 mL centrifuge tube to which two volumes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com