RPA primer and method for detecting schistosoma japonicum katsurada
A technology for schistosomiasis and amplification products, which is applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve problems such as difficulties in schistosomiasis prevention work, and achieve the effects of high sensitivity, good application prospects and convenient operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
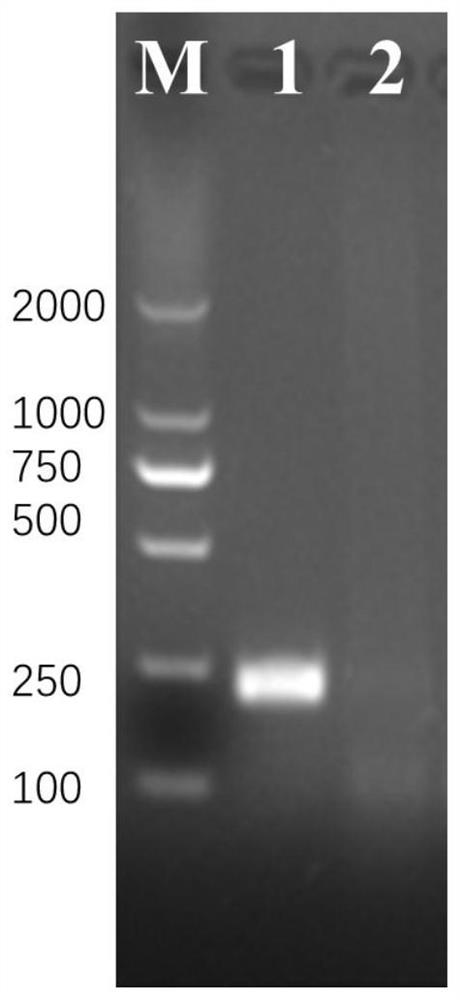
[0023] Example 1 RPA Detection of Schistosoma japonicum Adult Samples
[0024] 1) Design RPA primers: select the Sj28S ribosomal gene as the target gene, and design specific primers. Firstly, conduct a large number of sequence analysis and alignment, and then borrow Primer Premier5 software, combined with manual design, and use experimental comparisons to screen out Optimal pair of primers:
[0025] RPA-F: 5'-CTTCGGTTAGTCTGCTGATGGCTTGTTTATG-3' (SEQ ID NO.1)
[0026] RPA-R:5'-CCTTAGTTCGGACTGAAAGCCGAACCTTTACC-3' (SEQ ID NO.2)
[0027] The specific amplification fragment corresponding to the above primers is located between the 851-1066 sites of the reference gene (GenbankZ46504.4) of Schistosoma japonicum, with a size of 216bp, and its nucleotide sequence is:
[0028] 5'-CTTCGGTTAGTCTGCTGATGGCTTGTTTATGTCACGTTGGCAGTTGCGTATGTGAGCTTTTGAATGGGCCAATAGTCTGTGGTGTAGTGGTAGACGATCCACCTGACCCGTCTTGAAACACGGAACCAAGGAGTTTAACATGTGCGCGAGTCATTGGGCGTTACGAAACCCAAAGGCGAAGTGAAGGTAAAGGTTCGGCTTTCAGTCCG...
Embodiment 2
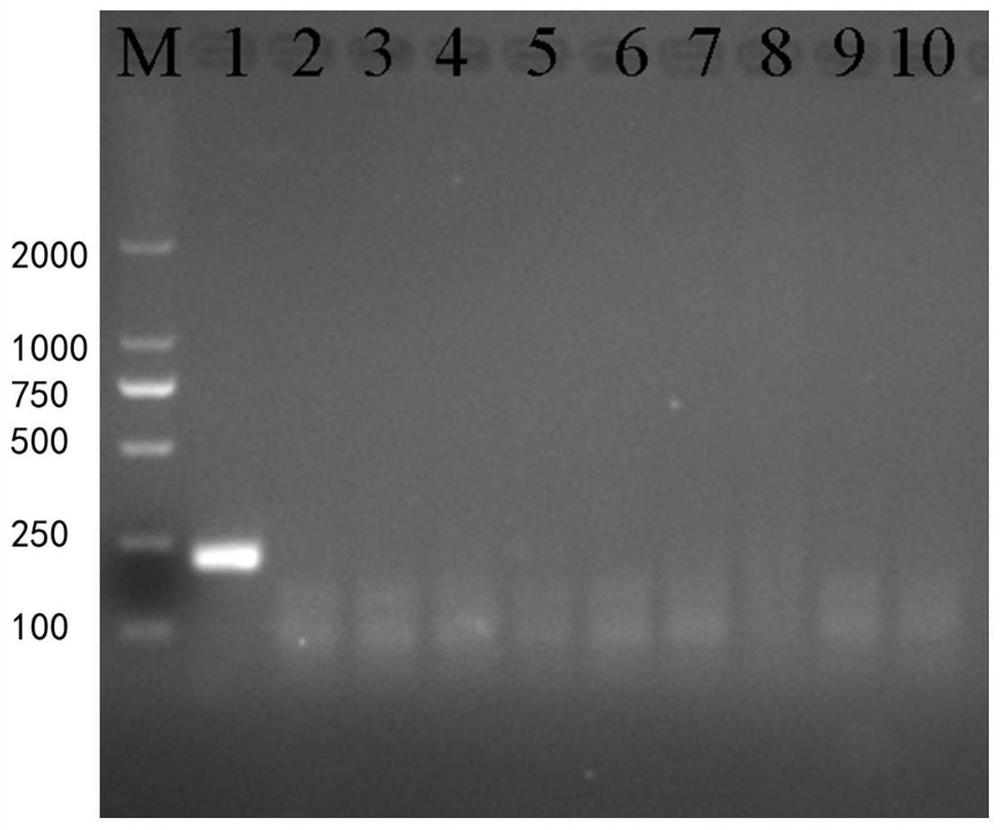
[0036] Example 2 RPA detection method specificity verification
[0037] Genomic DNA of Schistosoma japonicum, Schistosoma mansoni, Schistosoma haematobium, monofurcation cercariae, Clonorchis sinensis, Fasciola gigantea, Paragonimus guardis, Taenia saginata and negative snails were used as templates, and the negative control was sterilized double distilled water , carry out RPA reaction according to the method for embodiment 1, and the amplified product is detected, the results are shown in figure 2 .
[0038] figure 2It shows that only the swimming lane added with Schistosoma japonicum genomic DNA has obvious amplified bands, and the others have no any amplified bands, indicating that the primers of the present invention have high specificity for Schistosoma japonicum, and the RPA detection method has better specificity.
Embodiment 3
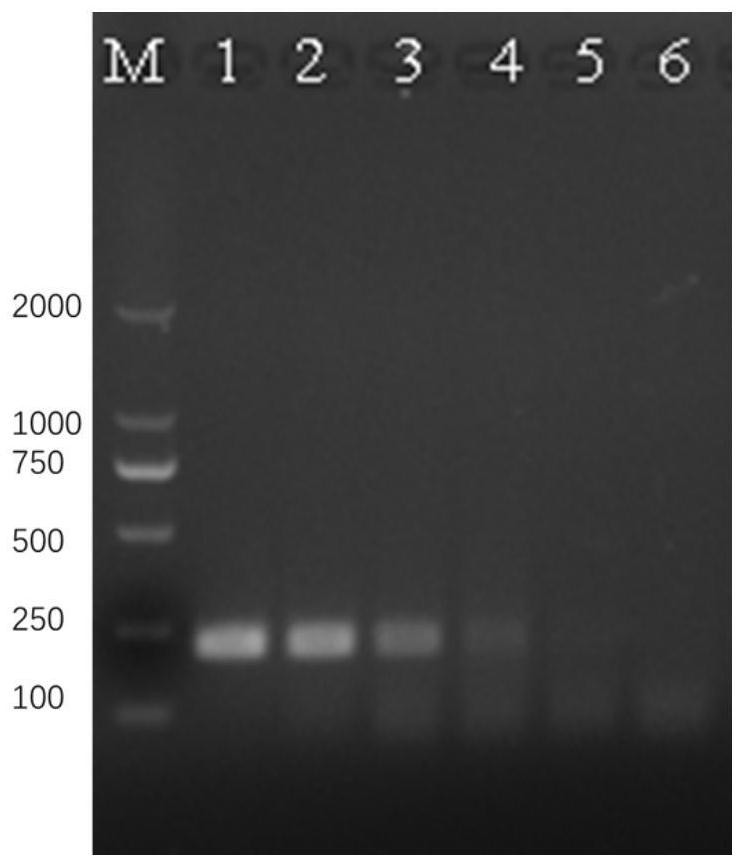
[0039] Example 3 RPA detection method sensitivity verification
[0040] 1) Select the total DNA sample of Schistosoma japonicum adult worms to be serially diluted 10 times, select 5 concentration gradients: 100, 10, 1pg / μL, 100, 10fg / μL, and the negative control is sterilized double distilled water, using the method in Example 1 test, see the results image 3 .
[0041] The results show that when the sample concentration is greater than or equal to 100fg / μL, the obvious target band can be amplified; when the sample concentration is 10fg / μL, the corresponding target band can no longer be amplified; the negative control has no amplified band band, indicating that the experimental process was free of pollution. The results showed that the minimum detectable concentration of RPA reaction to the genomic DNA of adult Schistosoma japonicum was 100fg / μL.
[0042] 2) Construct the 472bp target fragment including the sequence of SEQ ID NO.3 on the Puc57-T vector, and extract the reco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com