Amplification-free RNA quantitative detection method
A quantitative detection method and content technology, applied in the field of molecular biological detection, can solve problems such as complex environmental background, short length of RNA fragments, and high sequence similarity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1, expression and purification of LbuCas13a protein
[0036]First, 100 mg / μL of ampicillin (Shanghai Shengong) was added to the rich medium (Terrific Borth growth media, OXOID). Under the condition of 37°C, the Escherichia coli Rosetta (DE3) constructed with pET28a-His6-SUMO-LbuCas13a plasmid was cultivated in this medium until the bacterial density reached OD (600nm) of 0.6. Then, add isopropyl-1-thio-β-D-galactopyranose (isopropyl-1-thio-b-D-galactopyranoside, purchased from sigma) with a final concentration of 100 μM to the above bacterial culture solution, and lower the temperature to Continue culturing at 26°C for 4 hours to induce protein expression. Next, the bacteria cultured above were collected and ultrasonically lysed after adding a lysis buffer containing 20 mM Tris-HCl, 1 M NaCl, 10% glycerol and pH 7.5. Then the bacterial lysate was centrifuged at 4° C. at a speed of 8000 rpm, and the obtained supernatant was incubated with a nickel-containing...
Embodiment 2
[0037] Example 2, single-molecule RNA detection capability verification
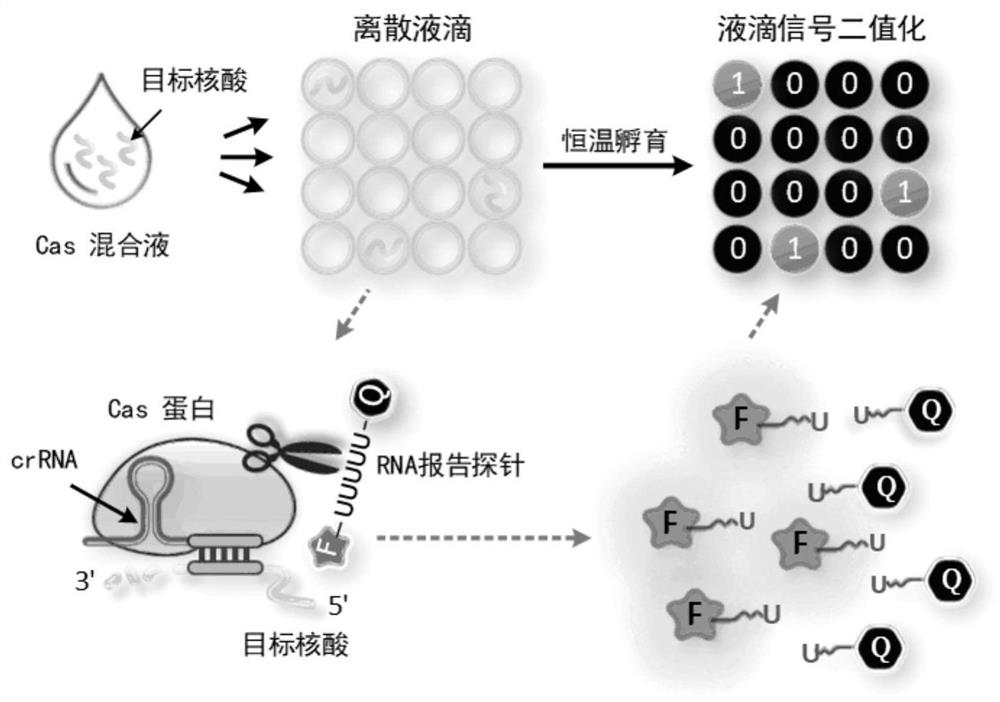
[0038] In order to verify the feasibility of single-molecule RNA detection by the technical solution provided by the present invention, the verification experimental operation steps are as follows:
[0039] (1) Design crRNA-miR-17 (as shown in sequence 2 of the sequence listing) for the microRNA-17 sequence (as shown in sequence 1 of the sequence listing): GACCACCCCAAAAAUGAAGG GGACUAAAAC CCUGCACUGUAAGCACUUU G, the nucleotide sequence shown underlined is complementary to the microRNA-17 sequence.
[0040] (2) Prepare 9 μL of Cas13 reaction system, whose components mainly include 20 nM LbuCas13a protein prepared in Example 1, 10 nM crRNA-miR-17, 300 nM FQ 5U RNA fluorescent reporter probe (FAM-UUUUU-BHQ1) and 1× reaction Buffer, wherein the reaction buffer contains 10mM Tris-HCl, 1.5mM MgCl 2 , 50mM KCl, solution pH 8.9.
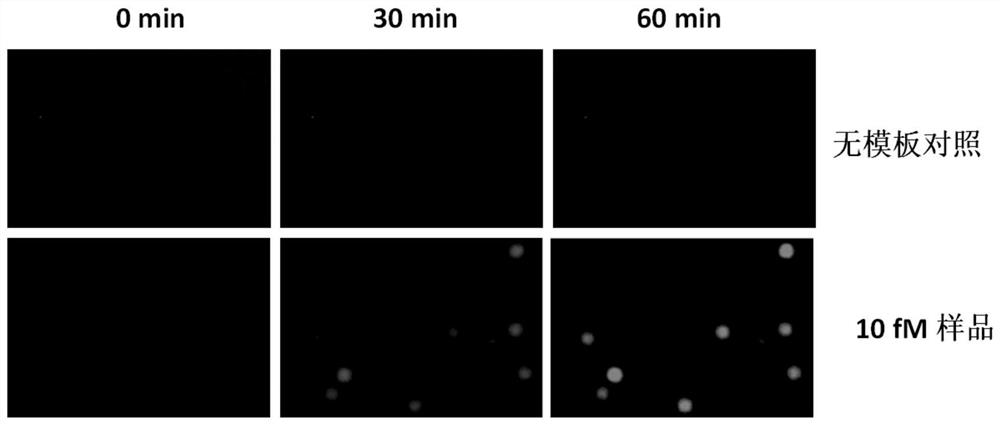
[0041] (3) 1 μL of microRNA-17 with a concentration of 100 fM was used as the ta...
Embodiment 3
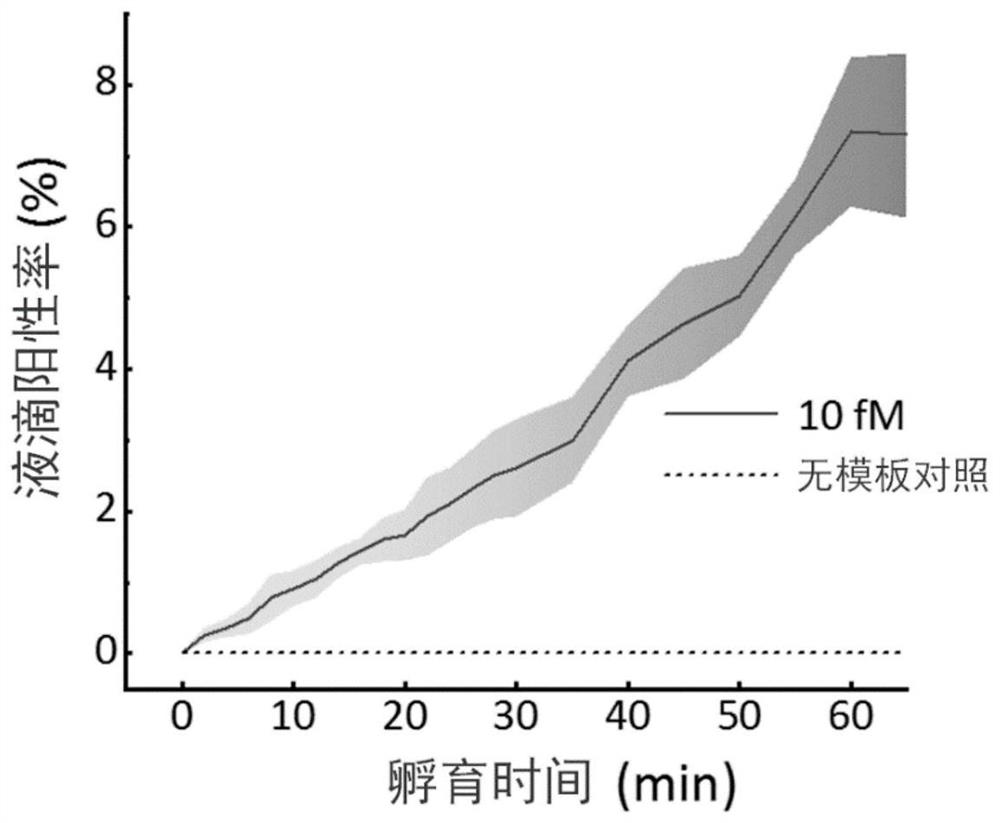
[0047] Example 3, using gradient dilution micro RNA-17 samples to verify the quantitative detection ability
[0048] (1) Dilute the known concentration of microRNA-17 synthetic target (purchased from Treasure Bioengineering (Dalian) Co., Ltd.) 10-fold gradient to prepare quantitative standards with concentrations of 1pM, 100fM, 10fM, 1fM, 100aM, and 10aM, respectively , while DEPC water without template was used as a negative control.
[0049] (2) Prepare a reaction system whose components mainly include 20nM LbuCas13a protein prepared as in Example 1, 10nMcrRNA-miR-17, 300nM FQ 5U RNA fluorescent reporter probe (FAM-UUUUU-BHQ1) and 1× reaction buffer, Wherein the reaction buffer contains 10mM Tris-HCl, 1.5mM MgCl 2 , 50mM KCl, solution pH 8.9.
[0050] (3) Mix the quantitative standard in step (1) with the reaction system in step (2) above to form a large number of water-in-oil discrete micro-droplets with a diameter of 30 μm, and the number of micro-droplets is not less th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com