New application of PTX-3 in medical detection
A PTX-3, 1. PTX-3 technology, applied in the field of biomedicine, can solve the problems of poor prognosis, recurrence, sensitivity to chemotherapeutic drugs, etc., and achieve good clinical application value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1: Preliminary screening of differentially expressed genes in ovarian cancer
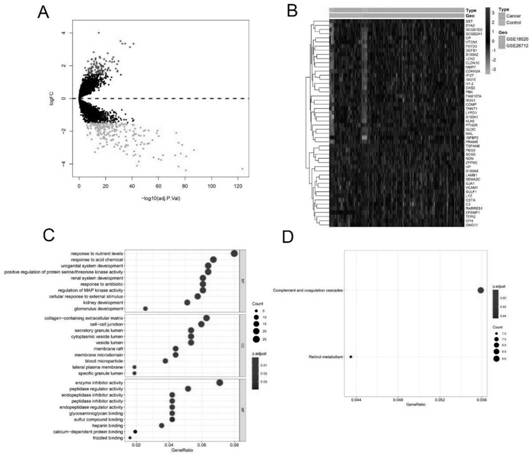
[0058] Two ovarian cancer datasets were selected using the GEO database: GSE18520 and GSE26712. The GSE18520 dataset is based on the GPL570 platform (HG-U133_Plus_2; Affymetrix Human Genome U133 Plus 2.0 Array), which contains 53 high-grade serous papillary carcinoma samples and 10 paracancerous carcinoma samples. The GSE26712 dataset is based on the GPL96 platform (HG-U133A; Affymetrix Human Genome U133A Array), which contains 10 normal ovarian epithelial samples and 185 primary ovarian cancer samples. By using the R package "limma", the differences between 20 normal ovarian samples and 238 ovarian cancer samples were analyzed. |logFC|>1.5, and p-value<0.05 were defined as differentially expressed genes. A total of 100 up-regulated genes and 228 down-regulated genes were analyzed by Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways by using t...
Embodiment 2
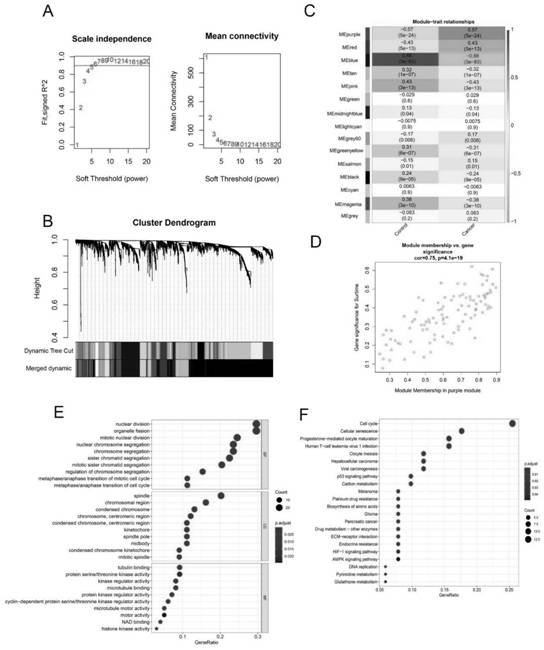
[0060] Example 2: Construction of weighted co-expression network for further screening of differentially expressed genes in ovarian cancer
[0061] The WGCNA software package was used to construct the gene co-expression network. The top 25% of genes in the variance plot were selected to construct a weighted co-expression network. The network modules are subdivided using the dynamic pruning tree algorithm. To test the stability of each identified module, train and test sets were randomly generated using Preserve Function Module Stability in the WGCNA package. Key modules were searched for correlations between modules and clinical characteristics assessed by the Pearson correlation test. Clinical features of the samples included normal tissue and ovarian cancer, and correlations between modules and features were calculated. Modules positively associated with ovarian cancer are thought to play a role in the pathogenesis of the disease. On the other hand, genes in modules posi...
Embodiment 3
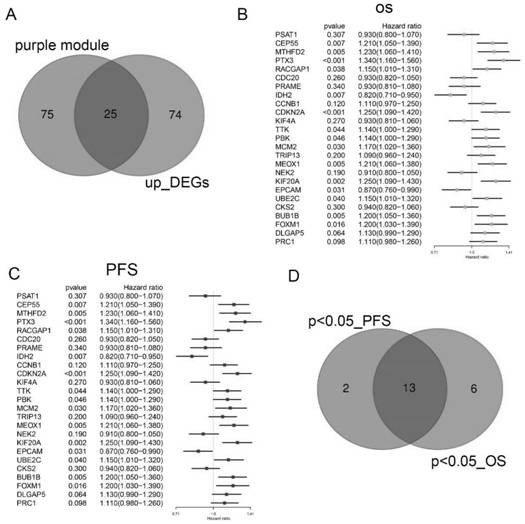
[0063] Example 3: Survival analysis of differentially expressed genes in ovarian cancer
[0064] After combining differentially expressed genes and modular genes, we selected overlapping genes as central genes ( image 3 A). Ovarian cancer data from the Kaplan Meier-Plotter database (http: / / kmplot.com / analysis / ) were used for survival analysis of central genes. The OS (overall survival) forest plot of these genes is shown in image 3 As shown in B, and the progression-free survival (PFS) forest plot of these genes is also shown in image 3 As shown in C, and the progression-free survival (PFS) forest plot of these genes is also shown in image 3 C shown. A total of 13 genes were identified, including BUB1B, KIF20A, MCM2, CEP55, MTHFD2, FOXM1, PBK, CDKN2A, PTX3, RACGAP1, MEOX1, UBE2C, and IDH2, which were associated with OS and PFS in ovarian cancer patients ( image 3 D).
[0065] Conclusion: BUB1B, KIF20A, MCM2, CEP55, MTHFD2, FOXM1, PBK, CDKN2A, PTX3, RACGAP1, MEOX1, U...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com