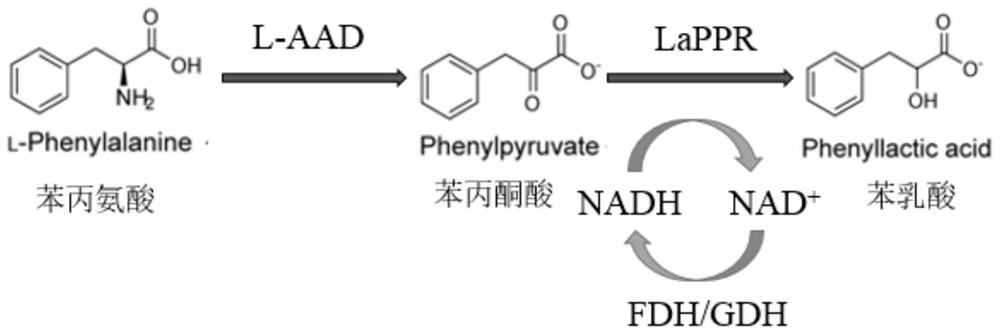
Method for synthesizing L-phenyllactic acid through whole-cell catalysis of recombinant microorganisms
A technology of phenyllactic acid and host cells, applied in the field of enzyme engineering and genetic engineering, can solve the problems of high cost and pollution of phenyllactic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
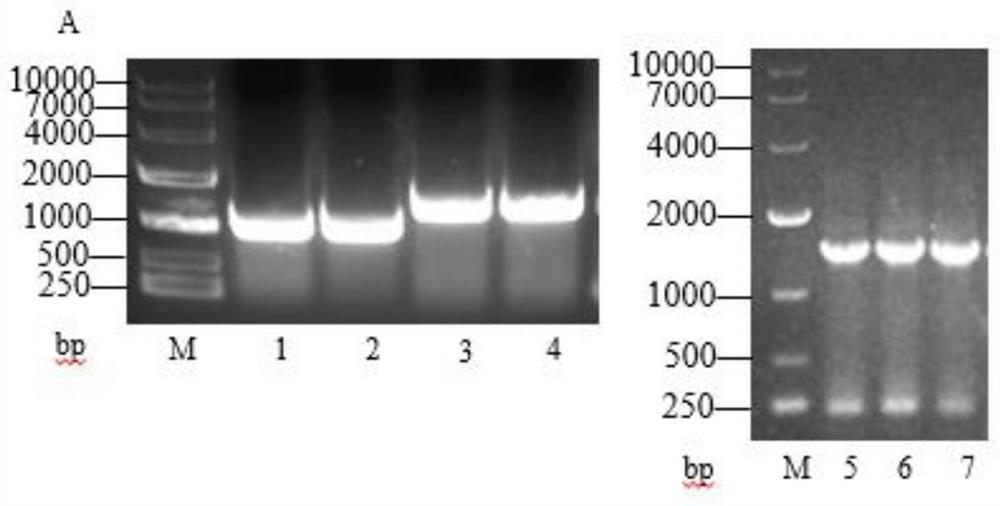
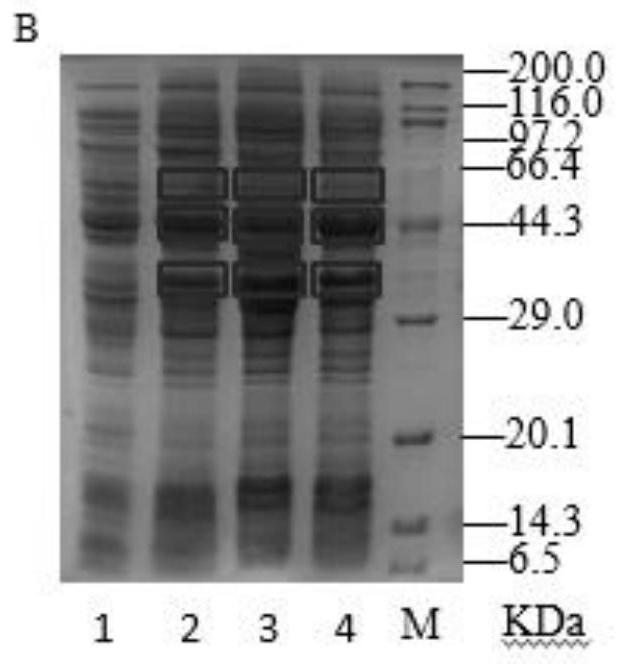
[0066] Embodiment 1: Construction of phenylpyruvate reductase expression vector
[0067] Specific steps are as follows:
[0068] Using the Lactobacillus sp.CGMCC 9967 genome as a template, the phenylpyruvate reductase gene lappr as shown in SEQ ID NO.5 was amplified, and it was combined with the pET-28a(+) expression plasmid through the restriction endonuclease HindⅢ enzyme Ligation was performed after cutting to obtain the recombinant plasmid pET28a-lappr.
[0069] The primer sequences used to construct the expression vector are as follows:
[0070] Upstream primer (including HindⅢ restriction site): AATTCGAGCTCCGTCGAC AAGCTT GATGGGCAGCAGCCATCA (SEQ ID NO. 7);
[0071] Downstream primer (including HindⅢ restriction site): GGTGCTCGAGTGCGGCCGC AAGCTT TTAATAGCCGCGGGTCAGAT (SEQ ID NO. 8).
Embodiment 2
[0072] Embodiment 2: Construction of glucose dehydrogenase expression vector
[0073] Specific steps are as follows:
[0074] Using the Bacillus subtilis genome as a template, the glucose dehydrogenase gene as shown in SEQ ID NO.6 was amplified, and it was combined with the pET28a-lappr expression plasmid constructed in Example 1 through the restriction endonuclease NcoI Ligation was performed after digestion to obtain the recombinant plasmid pET28a-lappr-gdh.
[0075] The primer sequences used to construct the expression vector are as follows:
[0076] Upstream primer (including NcoⅠ restriction site): ACTTTAAGAAGGAGATATA CCATGG ATGGGTTATCCGGATTTAAAAGGAAAAG (SEQ ID NO. 9);
[0077] Downstream primer (including NcoⅠ restriction site): TGATGATGATGATGATGGCTGCTGC CCATGG TTAACCGCGGC (SEQ ID NO. 10).
Embodiment 3
[0078] Embodiment 3: Construction of L-amino acid deaminase expression vector
[0079] Specific steps are as follows:
[0080] Using the Proteus vulgaris genome as a template, the L-amino acid deaminase gene as shown in SEQ ID NO.4 was amplified, and it was restricted with the pET28a-lappr-gdh expression plasmid constructed in Example 2. Endonuclease XhoI digested and ligated to obtain recombinant plasmid pET28a-lappr-gdh-laad.
[0081] The primer sequences used to construct the expression vector are as follows:
[0082] Upstream primer (including XhoⅠ restriction site): AAGCTTGCGGCCGCA CTCGAG TAATACGACTCACTATAGGGGAATTGTGAG (SEQ ID NO. 11);
[0083] Downstream primer (including XhoI restriction site): GGTGGTGGTGGTG CTCGAG TTAAAAACGATATAAAGAAAACGGCTTCGGG (SEQ ID NO. 12).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com