Ultrahigh-sensitivity protease digital detection method
A digital detection and protease technology, applied in the field of enzyme detection, can solve the problems of limited sensitivity and false negative of protease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
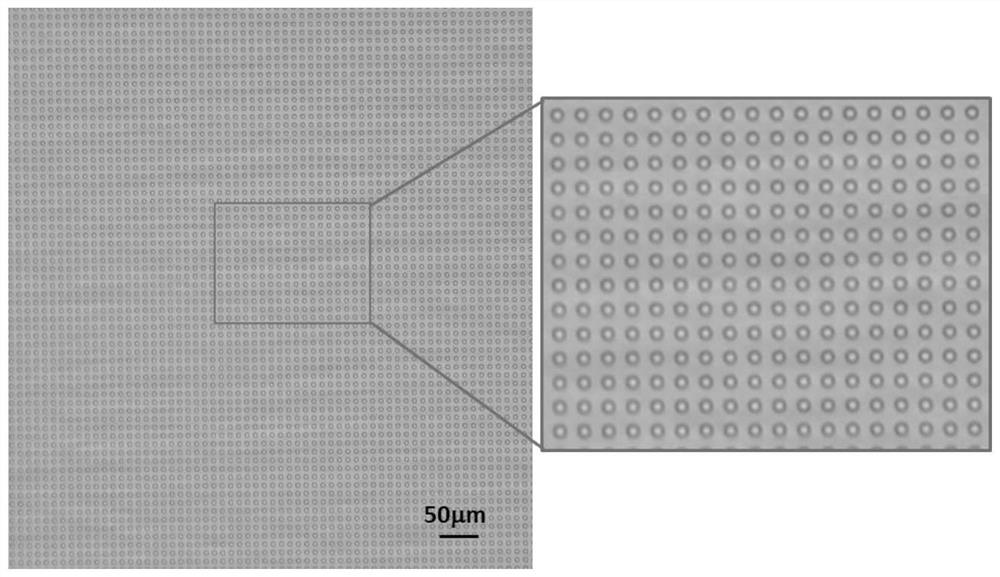
[0073] In this embodiment, a microarray microfluidic chip is prepared, and the chip consists of 2014 3D CAD software design, consisting of a microarray layer and a cover sheet layer, the microarray layer contains 1 million circular reaction microwells, each microwell can only accommodate one magnetic bead, and the volume of the microwell is fL level, the microarray The material of the layer and the cover sheet layer is a mixed silica gel (polydimethylsiloxane (PDMS) and tetraethyl orthosilicate, the mass ratio is 10:1), which is produced by injection molding process.
[0074] The specific production process includes:
[0075] (1) Weigh PDMS and tetraethyl orthosilicate according to the mass ratio of 10:1, after fully stirring and mixing, carry out degassing and defoaming treatment in a vacuum device;
[0076] (2) Inject the degassed and defoamed mixture into the microarray layer mold, and cure at 80° C. for 30 minutes;
[0077] (3) peel off the formed silica gel chip from t...
Embodiment 2
[0082] This embodiment prepares the magnetic bead-antibody complex, and the preparation method of the magnetic bead-antibody complex comprises the following steps:
[0083] (1) Take 50 μL carboxy-modified magnetic beads (5 mg / mL), wash them twice with PBS, add pre-cooled 20 μL EDC (20 mg / mL) and 20 μL NHS (20 mg / mL) respectively, and use MES buffer for EDC and NHS Dissolve (20mM, pH 6), shake at room temperature for 30 minutes to activate the carboxyl group, and use a magnetic rack to enrich the magnetic beads;
[0084] (2) Use pre-cooled MES buffer to wash the magnetic beads enriched in step (1) 3 times, add 20 μg anti-3CL protease antibody, shake and react at room temperature for 2 hours, use a magnetic rack to enrich the magnetic beads, and discard the supernatant , wash the magnetic beads 3 times with pre-cooled MES buffer, add Tris-BSA buffer (25mM Tris-HCl, 0.1% Tween-20, 0.5% BSA, pH 7.4), shake at room temperature for 30min, and block the excess activated carboxyl grou...
Embodiment 3
[0086] This embodiment takes the 3CL protease of coronavirus as an example to detect.
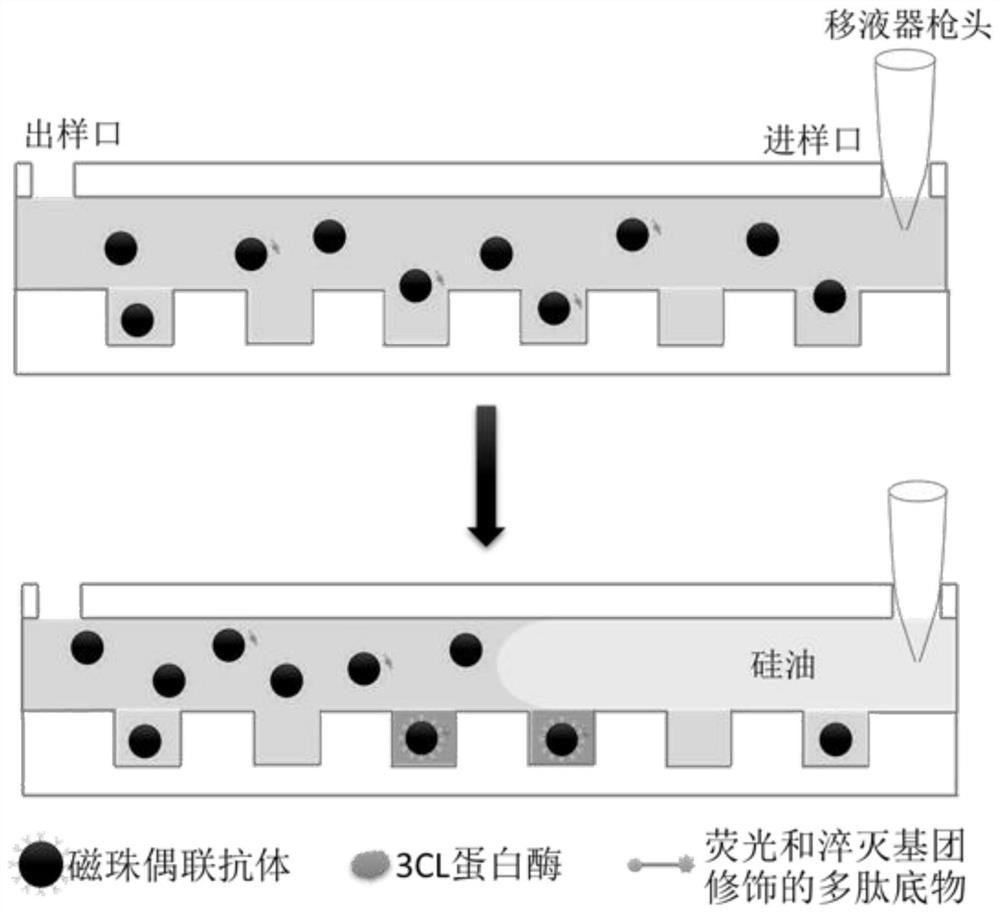
[0087] The schematic diagram of the detection is as figure 2 As shown, it specifically includes the following steps: (1) react the magnetic bead-antibody complex prepared in Example 2 with 3CL protease in PBS buffer at 4°C for 60 minutes to form a magnetic bead / antibody / 3CL protease complex, and enrich the Magnetic beads, discard the supernatant, add pre-cooled PBS to wash 3 times, and use Tris-HCl buffer (20mM, pH7.0) to resuspend;
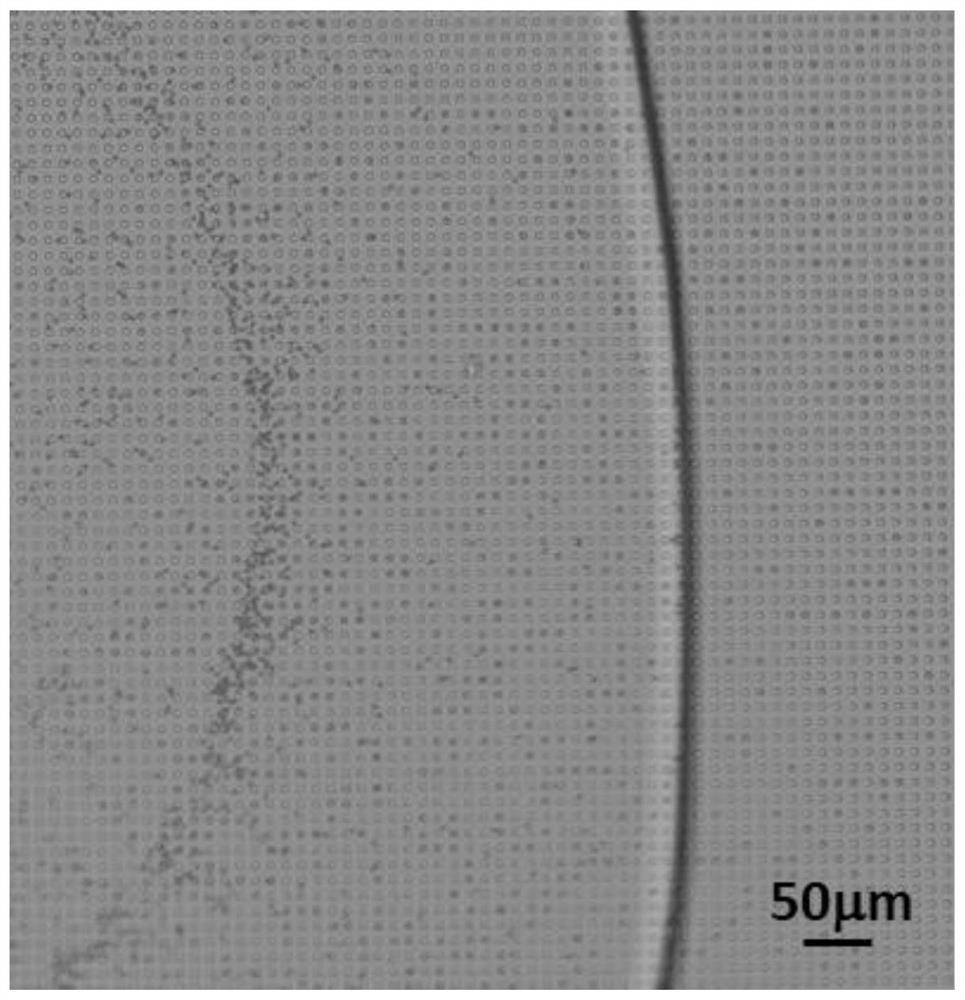
[0088] (2) Add fluorescent polypeptide substrate (sequence is Dabcyl-TSAVLQSGFRVM-FITC, wherein Dabcyl is fluorescent quenching group succinimidyl ester; FITC is fluorescein isothiocyanate) to step (1) resuspension liquid The final volume is 50 μL, and then the liquid is injected from the injection port of the microarray microfluidic chip prepared in Example 1. After standing at room temperature for 5 minutes, fluorine oil is added from the injection port, a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| recovery rate | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com