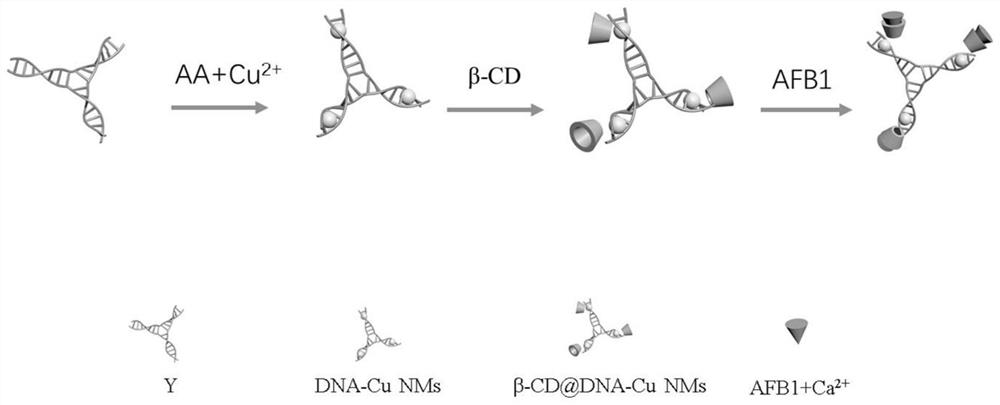
A method for detecting aflatoxin b1 based on fluorescent copper nanoparticles
An aflatoxin and nanoparticle technology, which is applied in the fields of analytical chemistry, materials science, and nano-biosensing to achieve highly sensitive fluorescence detection and high selectivity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] A method for preparing fluorescent copper nanoparticles β-CD@DNA-Cu NMs, comprising the steps of:
[0074] (1) Preparation of template strand Y-type complementary nucleic acid duplex (Y-type DNA):
[0075] Three equal oligonucleotide strands (Y 0a ,Y 0b ,Y 0c ) (Table 1) in MOPS buffer (10mM, pH 7.5, 150mMNaAc, 1mM MgCl 2 ) to obtain a mixed solution; then, the mixed solution was heated to 90°C, denatured for 5 minutes, and then slowly cooled to room temperature to obtain Y-shaped DNA; the Y-shaped DNA solution was stored in a -18°C refrigerator.
[0076] Table 1 DNA sequence of oligonucleotide chain
[0077] serial number sequence (from 5' to 3')) Y 0a
CCTGTCTGCCTAATGTGCGTCGTAAG SEQ ID NO.1 Y 0b
CTTACGACGCACAAGGAGATCATGAG SEQ ID NO.2 Y 0c
CTCATGATCTCCTTTAGGCAGACAGG SEQ ID NO.3
[0078] (2) Preparation of DNA-Cu NMs:
[0079] Mix Y-shaped DNA solution (500 μL, 2 μM) with ascorbic acid solution (500 μL, 1.25 mM), t...
Embodiment 2
[0083] The optimization of embodiment 2 fluorescent copper nanoparticles preparation technology
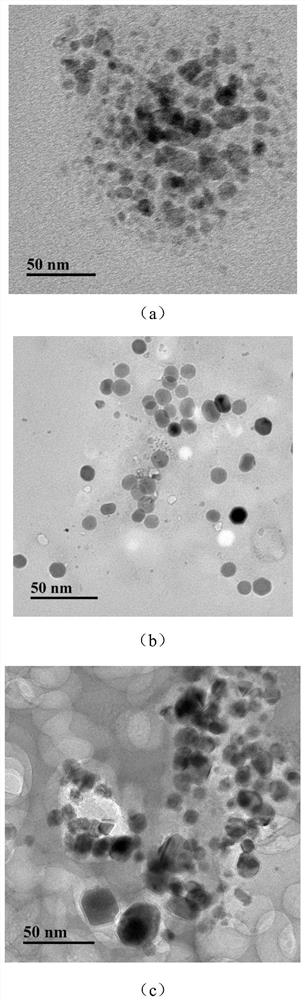
[0084] The concentration of the β-CD solution in Example 1 was adjusted to 0.5, 2, and 5 mM, and the others were kept the same as in Example 1 to obtain β-CD@DNA-Cu NMs.
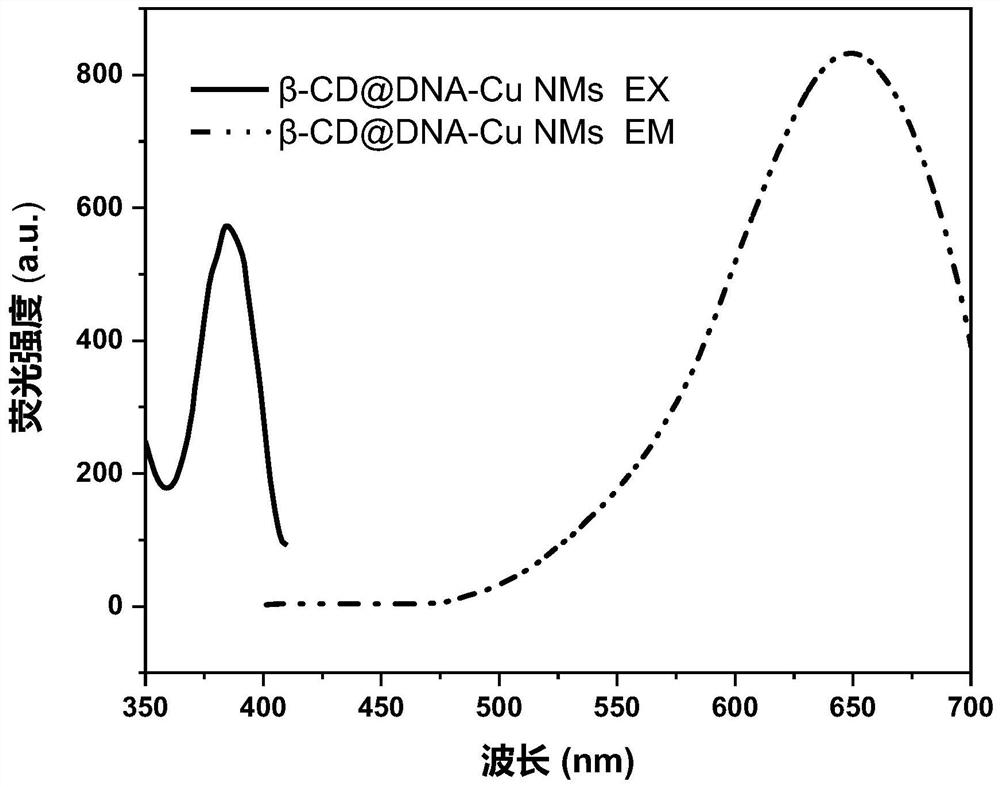
[0085] The obtained β-CD@DNA-Cu NMs were subjected to a fluorescence test. The conditions of the fluorescence test were: the excitation broadband was 10nm, the slit width was 10nm, and the excitation wavelength was 365nm; the test results were as follows:
[0086] figure 2 are the TEM images of β-CD@DNA-Cu NMs modified with different concentrations of β-CD; where (a) is 0.5mM, (b) is 2mM, and (c) is 5mM. From figure 2 It can be seen that as the concentration of β-CD increases from 0.5 to 2 and 5 mM, the average size of β-CD@DNA-Cu NMs increases from 10 nm to 12 nm and 14 nm, respectively. The concentration of β-CD is in the range of 0-5mM. With the increase of its concentration, the fluorescence is stronger...
Embodiment 3
[0089] A method for preparing a ratiometric fluorescence sensor of β-CD@DNA-Cu NMs-AFB1, comprising the steps of:
[0090] Add the β-CD@DNA-Cu NMs solution (100 μL, 200 μM) obtained in Example 1 into the AFB1 solution (900 μL, 10 ppb), shake at 400 rpm for 1 min, and mix well to obtain β-CD@DNA-Cu NMs-AFB1 ratiometric fluorescent sensor.
[0091] The obtained β-CD@DNA-Cu NMs-AFB1 ratiometric fluorescence sensor was tested for fluorescence. The test conditions were: the excitation broadband was 10nm, the slit width was 10nm, the excitation wavelength was 365nm, the detection was 433nm (AFB1) and 650nm (β- CD@DNA-Cu NMs) fluorescence values of the two emission peaks.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com