EGFR genotyping detection method based on nucleic acid tetrahedron probe modified printed gold electrode and kit
A tetrahedral probe and detection kit technology, used in biochemical equipment and methods, microbial determination/inspection, material analysis by electromagnetic means, etc., can solve the problems of insufficient sensitivity and low specificity, and improve the sensitivity , high sensitivity, less sample loading
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0029] The present invention will be further described in detail below in conjunction with the accompanying drawings and embodiments.
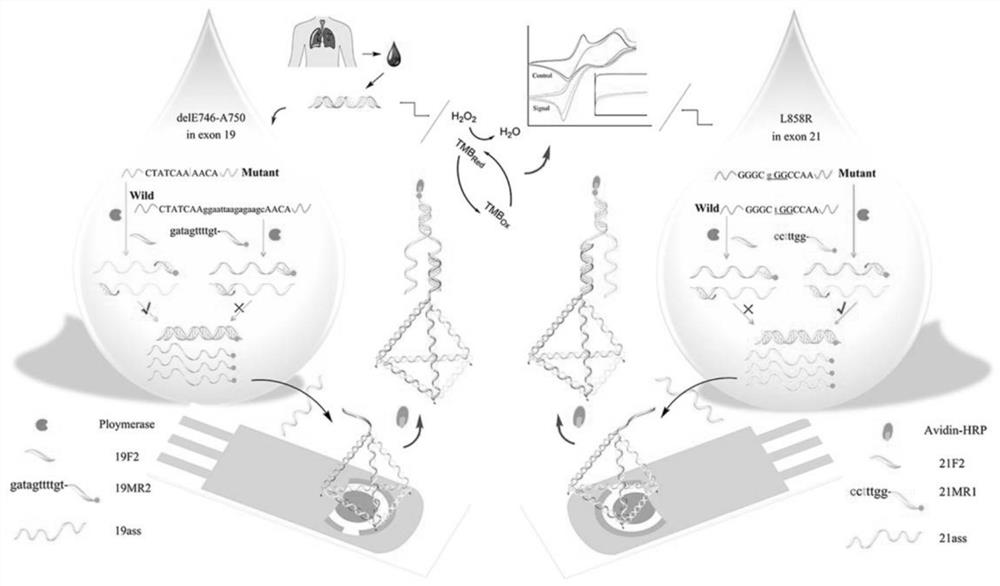
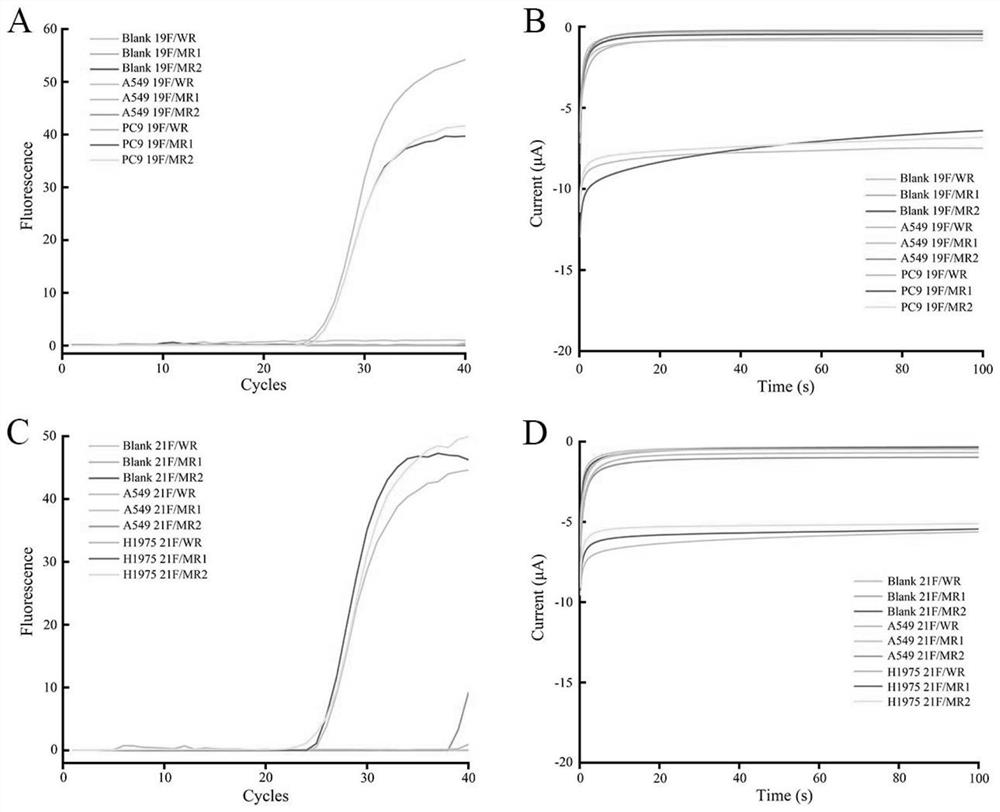
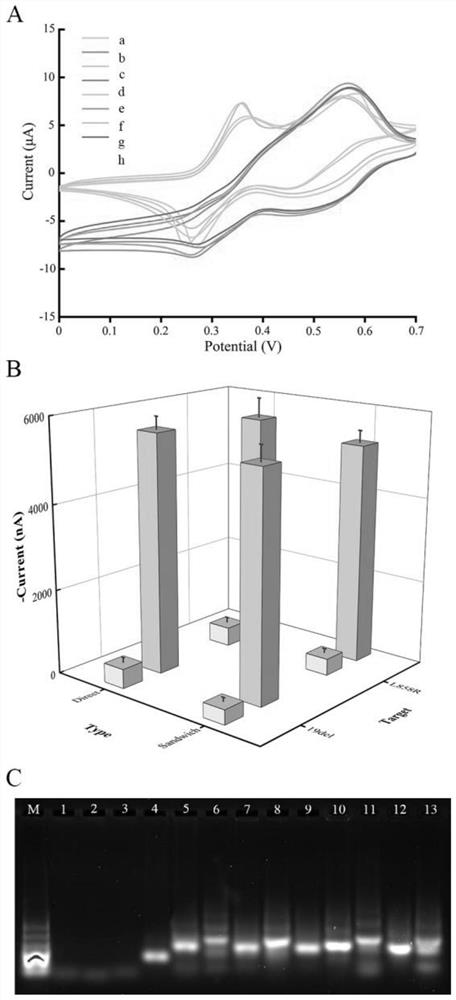
[0030] Such as Figure 1 to Figure 6 Shown, a kind of EGFR genotyping detection method based on nucleic acid tetrahedron probe modification printing gold electrode, comprises the following steps:
[0031] 1) Specific recognition and amplification of primers. In the reaction system, the concentration of the antisense primer chain is 200nM, the concentration of the sense primer chain is 20nM, and the thermal cycle conditions are 92°C for 15s, 58°C for 30s, 72°C for 30s, 45 cycles;
[0032] 2) Dissolve the four strands in TM hybridization buffer at equal concentrations, add TCEP at a final concentration of 30mM, heat the solution to 95°C for 5 minutes to fully untie the nucleic acid strands, then quickly transfer to 4°C for more than 30s, and the four nucleic acid strands The two are hybridized to form a tetrahedral structure, and the bottom thr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com