SNP (Single Nucleotide Polymorphism) molecular marker for improving disease resistance of procambarus clarkii and application
A technology of Crayfish and disease resistance, applied to SNP molecular markers and application fields for improving the disease resistance of Crayfish, can solve the problem of lack of transmission routes, difficult to define disease resistance traits, and disease resistance breeding of Crayfish. Slow progress and other problems, to achieve the effect of stable effect and excellent genotype disease resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
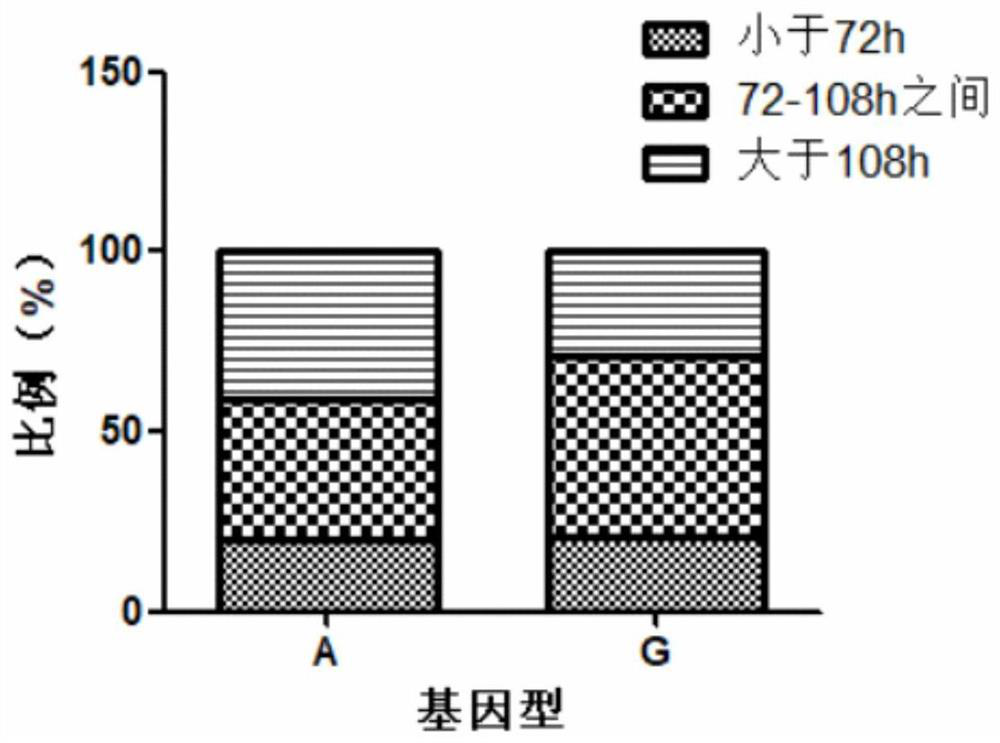
[0056] SNP site mining and disease resistance analysis of Crustin gene (GQ301202.1). A total of 176 Procambarus clarkii were selected for SNP site mining. A total of 90 Procambarus clarkii were selected for disease resistance analysis.
[0057] (1) Extract the hemolymph RNA of 176 individuals respectively, the specific method is as follows:
[0058] Use a 1ml syringe to extract 390-400μl (about 400μl) blood sample from the shrimp body, and mix it with ACD anticoagulant (formulation: 0.48g citric acid, 1.32g sodium citrate, 1.47g glucose, dissolved in 100ml double-distilled water) to 400μl, placed in EP tube, and placed on ice. Centrifuge at 800×g, 4°C for 20 minutes to separate blood cells; discard the supernatant, keep the white precipitate, add 200 μl of pre-cooled Trizol reagent, and use a grinder to grind until the white precipitate disappears, making the Trizol reagent solution pink, each sample Add 800μl Trizol reagent again; let it stand at room temperature for 5min,...
Embodiment 2
[0077] SNP site mining (screening) and disease resistance analysis for ALF gene (KU680792.1). A total of 176 Procambarus clarkii were selected for SNP site mining. A total of 90 Procambarus clarkii were selected for disease resistance analysis.
[0078] (1) Extract the hemolymph RNA of 176 Procambarus clarkii individuals respectively, and the specific steps are as follows:
[0079]390-400 μl (approximately 400 μl) blood sample was extracted from the shrimp of Procambarus clarkii using a 1ml syringe, and the volume ratio was 1:1 with ACD anticoagulant (0.48g citric acid, 1.32g sodium citrate, 1.47g glucose, dissolved in 100ml double distilled water) to 400μl, put in EP tube, and put on ice. Centrifuge at 800×g, 4°C for 20 minutes to separate blood cells; discard the supernatant, keep the white precipitate, add 200 μl of pre-cooled Trizol reagent, and use a grinder to grind until the white precipitate disappears, making the Trizol reagent pink. Add 800μl Trizol reagent; let s...
Embodiment 3
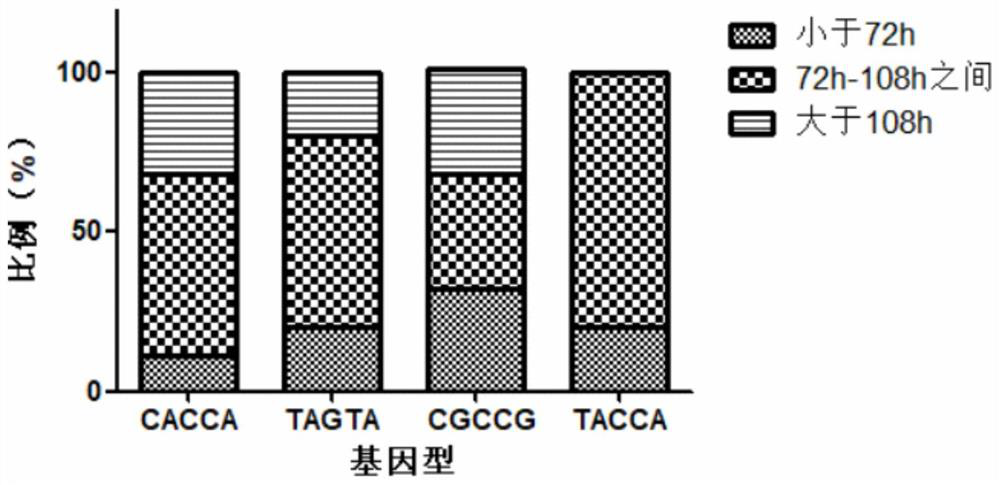
[0096] The Crustin gene and ALF gene were combined for SNP site mining and disease resistance analysis. A total of 176 Procambarus clarkii were selected for SNP site mining. A total of 90 Procambarus clarkii were selected for disease resistance analysis.
[0097] The Crustin gene and ALF gene were combined to further analyze the difference in disease resistance of different genotype combinations.
[0098] Specific steps are as follows:
[0099] (1) Extract the hemolymph RNA of 176 Procambarus clarkii individuals respectively, the specific method is as follows:
[0100] Use a 1ml syringe to extract 390-400μl (about 400μl) blood samples from Procambarus clarkii, and mix them with ACD anticoagulant (0.48g citric acid, 1.32g sodium citrate, 1.47g glucose, dissolved in 100ml double-distilled water) to 400μl, placed in EP tube, and placed on ice. Centrifuge at 800×g, 4°C for 20 minutes to separate blood cells; discard the supernatant, keep the white precipitate, add 200 μl of pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com