Method for synthesizing fludarabine phosphate through biological catalysis
A technology of fludarabine phosphate and catalyzed enzymes, applied in biochemical equipment and methods, botany equipment and methods, oxidoreductase, etc., can solve the problem of high cost, step-by-step operation of excessive materials, and preparation of enzyme-containing wet bacteria Complicated issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
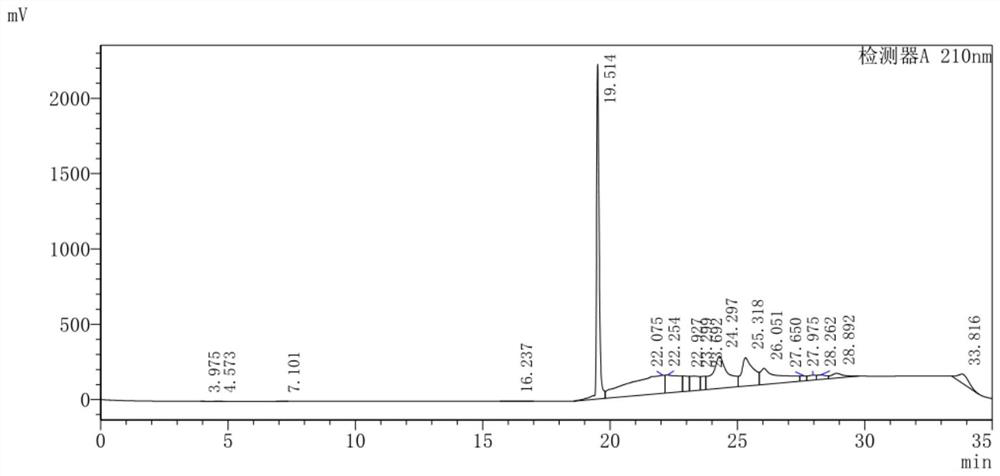
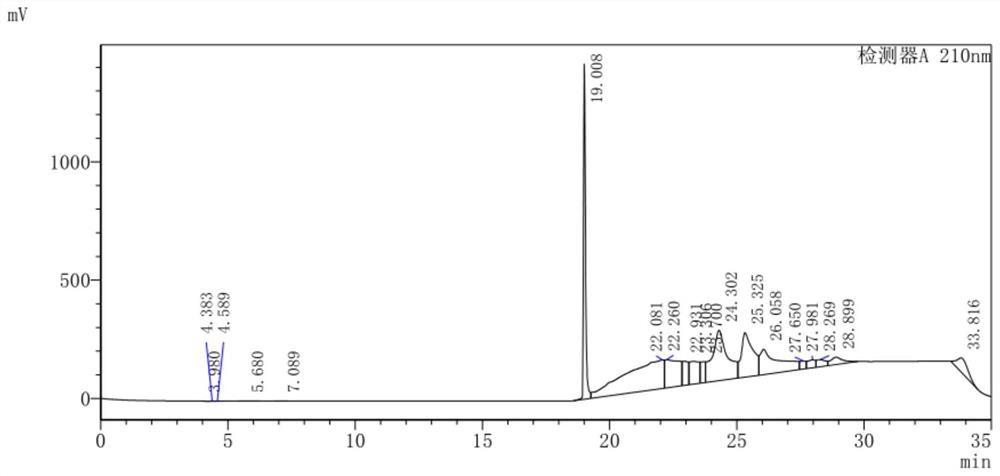
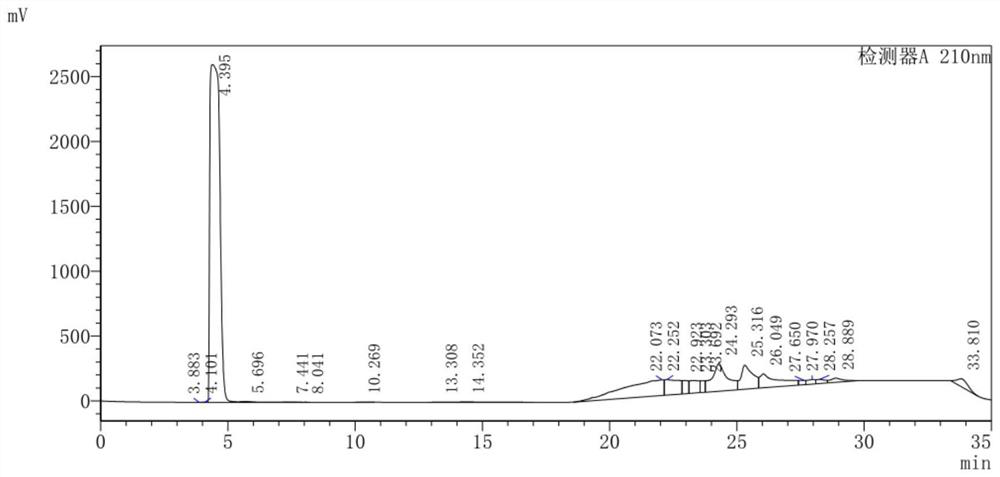
Image
Examples
Embodiment 1
[0027] The nucleotide sequence shown in the sequence list was selected to be ligated with the vector pBAD-D, and 20 μl of the ligated product was added to 200 μl of Escherichia coli TOP10 competent cells in ice bath, followed by ice bath for 30 min, heat shock at 42°C for 60 s, and ice bath for 5 min. Add 300 μl of 37°C non-antibiotic culture solution to the tube, repair it on a 200r shaker at 37°C for 1 hour, then spread it on an ampicillin-resistant solid LB plate and incubate at 37°C, and pick a single colony with an autoclaved toothpick after the colonies grow. First draw a line on the ampicillin-resistant plate for seed preservation, mark the corresponding bacteria and the line area on the plate, and then put the toothpick in the 20μl PCRMix system that has been added to the primer and mix it for a while. PCR amplification, PCR reaction conditions: 95°C, 15min, denaturation at 94°C for 15s, annealing at 55°C for 15s, extension at 72°C for 1min, 30 cycles, and finally 72°C,...
Embodiment 2
[0030] The nucleotide sequence shown in the sequence list was selected to be ligated with the carrier pBAD-D, and 20 μl of the ligated product was added to 200 μl of Escherichia coli TOP10 competent cells in ice bath, followed by ice bath for 30 min, heat shock at 42°C for 60 s, and ice bath for 5 min. Add 300 μl of 37°C non-antibiotic culture solution to the tube, repair it on a 200r shaker at 37°C for 1 hour, spread it on an ampicillin-resistant solid LB plate and incubate at 37°C, and pick a single colony with an autoclaved toothpick after the colonies grow. First draw a line on the ampicillin-resistant plate for seed preservation, mark the corresponding bacteria and the line area on the plate, and then put the toothpick in the 20μl PCR Mix system that has been added to the primer and stir it. Carry out PCR amplification, the PCR reaction conditions are: 95°C, 15min, denaturation at 94°C for 15s, annealing at 55°C for 15s, extension at 72°C for 1min, 30 cycles, and finally 7...
Embodiment 3
[0033] The nucleotide sequence shown in the sequence list was selected to be ligated with the carrier pBAD-D, and 20 μl of the ligated product was added to 200 μl of Escherichia coli TOP10 competent cells in ice bath, followed by ice bath for 30 min, heat shock at 42°C for 60 s, and ice bath for 5 min. Add 300 μl of 37°C non-antibiotic culture solution to the tube, repair it on a 200r shaker at 37°C for 1 hour, spread it on an ampicillin-resistant solid LB plate and incubate at 37°C, and pick a single colony with an autoclaved toothpick after the colonies grow. First draw a line on the ampicillin-resistant plate for seed preservation, mark the corresponding bacteria and the line area on the plate, and then put the toothpick in the 20μl PCR Mix system that has been added to the primer and stir it. Carry out PCR amplification, the PCR reaction conditions are: 95°C, 15min, denaturation at 94°C for 15s, annealing at 55°C for 15s, extension at 72°C for 1min, 30 cycles, and finally 7...
PUM
| Property | Measurement | Unit |
|---|---|---|
| decomposition temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com