Method for determining total number of viable bacteria based on WST-8 chromogenic reaction
A WST-8, color reaction technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, color/spectral characteristic measurement, etc., can solve problems such as low sensitivity, difficulty in color reaction, lack of applied research, etc. , to achieve high sensitivity, wide range of bacterial detection, and improved sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0051] A method for measuring the total number of live bacteria based on WST-8 color reaction, comprising the following steps:
[0052] (1) Make a standard curve
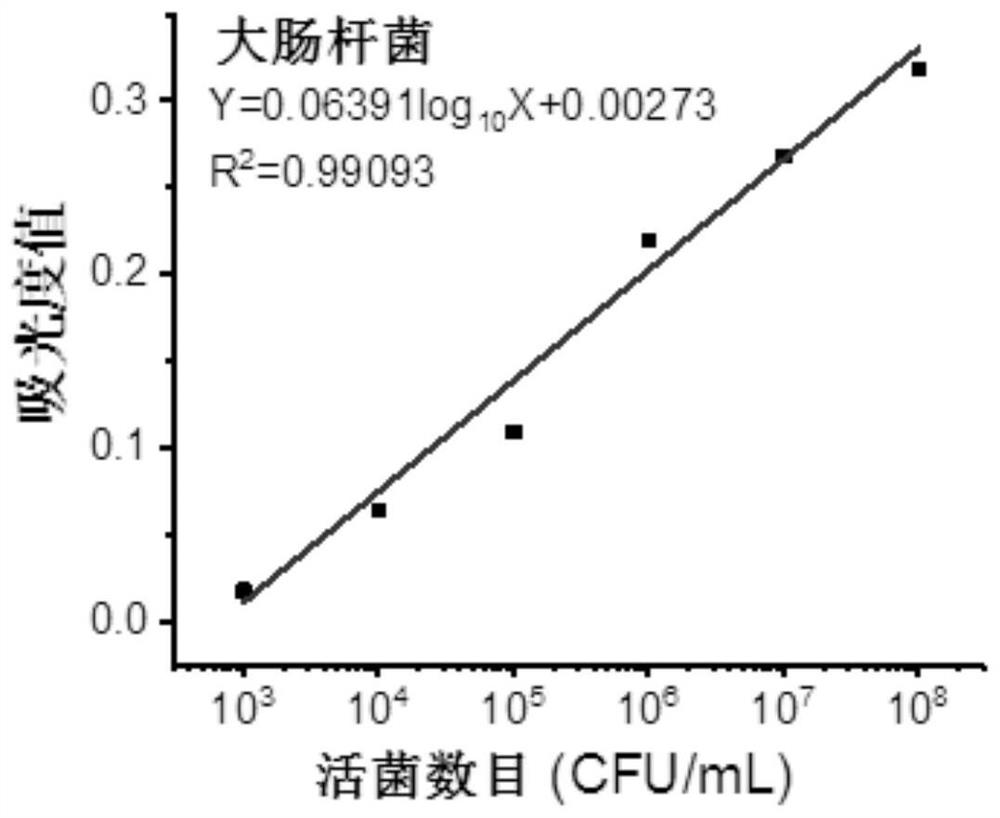
[0053] (11) Resuspend the Escherichia coli bacterial liquid with PBS buffer (pH=7.4) and carry out gradient dilution to prepare standard sample suspensions with different concentration gradients (for example, 10 3 CFU / mL, 10 4 CFU / mL, 10 5 CFU / mL, 10 6 CFU / mL, 10 7 CFU / mL, 10 8 CFU / mL);
[0054] (12) Prepare a chromogenic substrate solution comprising WST-8, electron carriers and nutrients, the concentration of WST-8 is 0.05mM, the concentration of electron carriers is 2μM, and the concentration of nutrients sodium chloride is 1mM;
[0055] Mix the standard sample suspensions with different concentration gradients with the chromogenic substrate solution respectively, and react for 2 hours to obtain the standard reaction solution to be tested;
[0056] (13) measure the absorbance value at 450nm of the standard...
Embodiment 2
[0065] A method for measuring the total number of live bacteria based on WST-8 color reaction, comprising the following steps:
[0066] (1) Make a standard curve
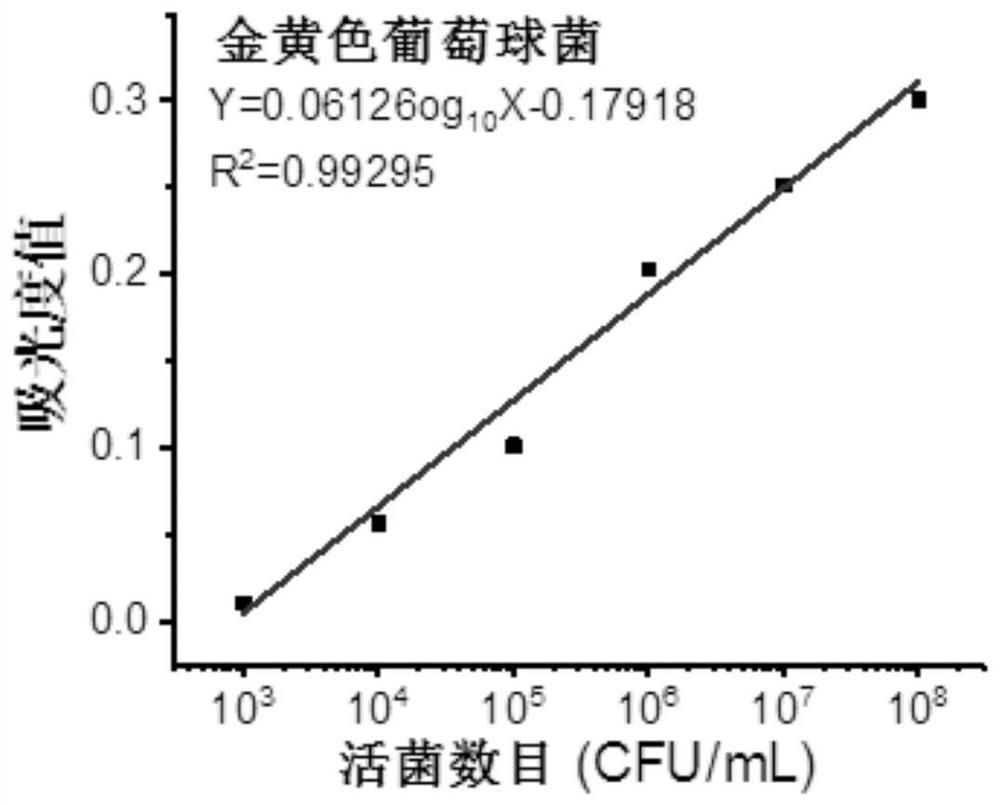
[0067] (11) Resuspend the Staphylococcus aureus bacterial liquid with PBS buffer (pH=7.4) and carry out gradient dilution to prepare standard sample suspensions with different concentration gradients (for example, 10 3 CFU / mL, 10 4 CFU / mL, 10 5 CFU / mL, 10 6 CFU / mL, 10 7 CFU / mL, 10 8 CFU / mL);
[0068] (12) Prepare a chromogenic substrate solution comprising WST-8, electron carriers and nutrients, the concentration of WST-8 is 0.05mM, the concentration of electron carriers is 2μM, and the concentration of nutrient peptone is 0.2mM;
[0069] Mix the standard sample suspensions with different concentration gradients with the chromogenic substrate solution respectively, and react for 2 hours to obtain the standard reaction solution to be tested;
[0070] (13) measure the absorbance value at 450nm of the standard r...
Embodiment 3
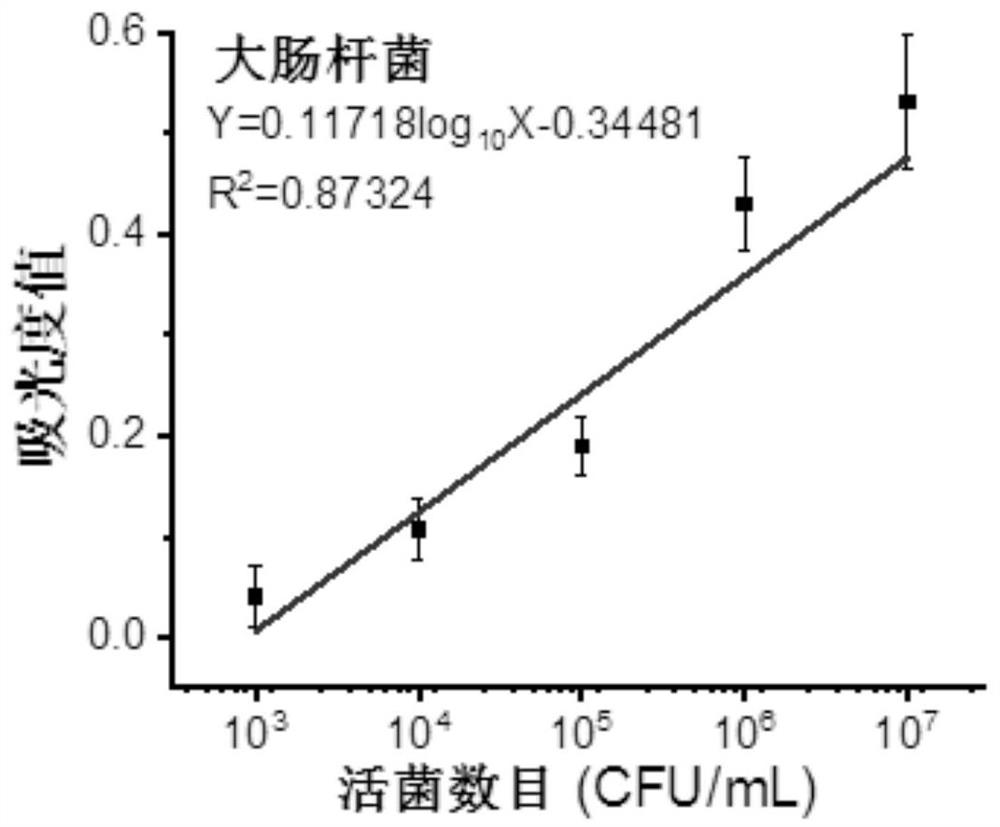
[0078] A method for determining the total number of viable bacteria based on the WST-8 color reaction, compared with Example 1, the difference is that the reaction time between Escherichia coli and the color substrate is 6h. Others are substantially the same as in Embodiment 1, and will not be repeated here.
[0079] The standard curve is Y=0.11718log 10 X-0.34481 (R 2 =0.87324), the linear concentration range of detectable bacteria is 10 3 -10 7 CFU / mL.
[0080] Adopt step (2) similar method to test unknown escherichia coli concentration, obtain escherichia coli concentration and be 8.9 * 10 7 CFU / mL; the concentration of Escherichia coli measured by plate counting method was 6.79×10 7 CFU / mL, the relative standard deviation of the bacterial concentration obtained by the two test methods is 31.08%. It can be seen that when the reaction time is too long, the bacterial growth enters the logarithmic phase and increases rapidly, which makes the linear relationship poor. The...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com