Polybrominated diphenyl ether sensory protein and whole-cell microbial sensor constructed by polybrominated diphenyl ether sensory protein
A microbial sensor, polybrominated diphenyl ether technology, applied in the biological field, can solve the problems of few biological components, low bioavailability, lack of discovery and research of biodegradation pathways, etc., and achieve the effect of improving monitoring sensitivity and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: Acquisition of polybrominated diphenyl ether recognition gene elements
[0023] Configure inorganic salt medium, and each liter of water contains the following substances (g / L): Na 2 HPO 4 12H 2 O 2.0, KH 2 PO 4 0.7, NH 4 Cl 0.5, NaCl 0.3, MgSO 4 ·7H 2 O 0.1, CaSO 4 2H 2 O 0.05, FeCl 3 ·6H 2 O 0.2×10 -3 、NaMoO 4 0.2×10 -3 , MnCl 2 4H 2 O 0.2×10 -3 , CuCl 2 2H 2 O 0.2×10 -3 , ZnSO 4 0.2×10 -3 、H 3 BO 3 0.3×10 -3 、CoCl 2 ·6H 2 O 0.4×10 -3 , peptone 0.2, yeast extract 1.0, glucose 5.0. Inoculate S. xenophagum hydrophobic strain C1 (=CCTCC AB 2015198=KCTC 42740) and hydrophilic strain C2 (=CCTCC AB2015427=KCTC 52051) into LB liquid medium, culture in a shaker at 200rpm / min at 30°C To logarithmic growth phase, cell OD 600 The value is about 1.0. Centrifuge at 10,000×g for 10 minutes, remove the supernatant, and collect the bacteria. After washing the cells twice with inorganic salt medium, resuspend the cells with a certain volume...
Embodiment 2
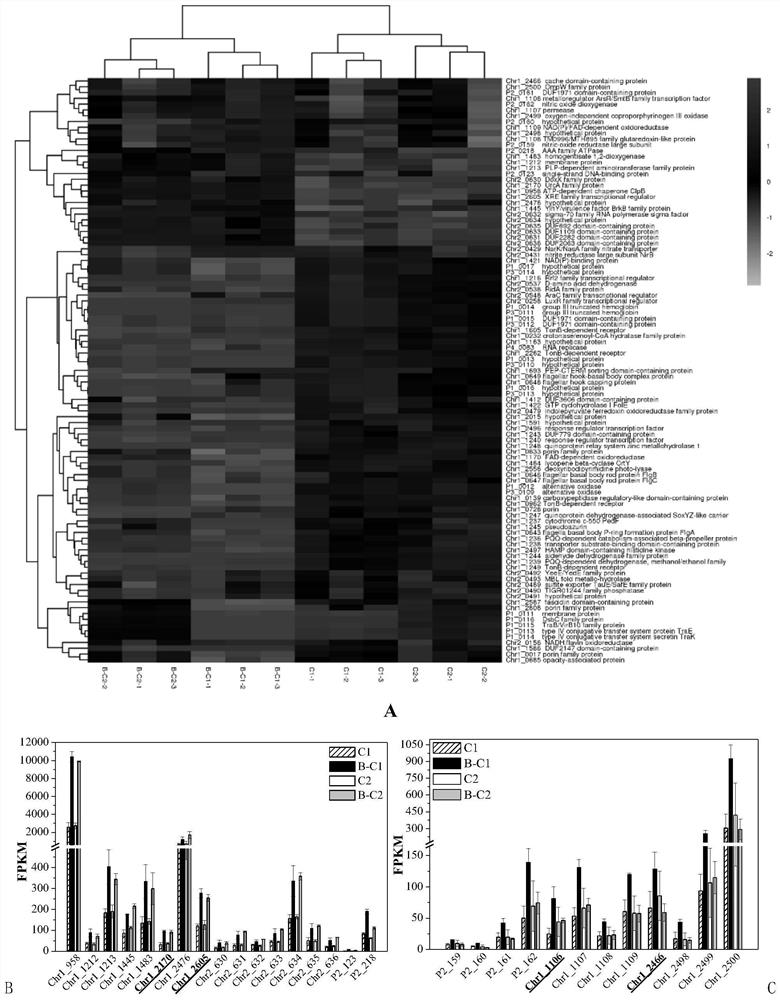
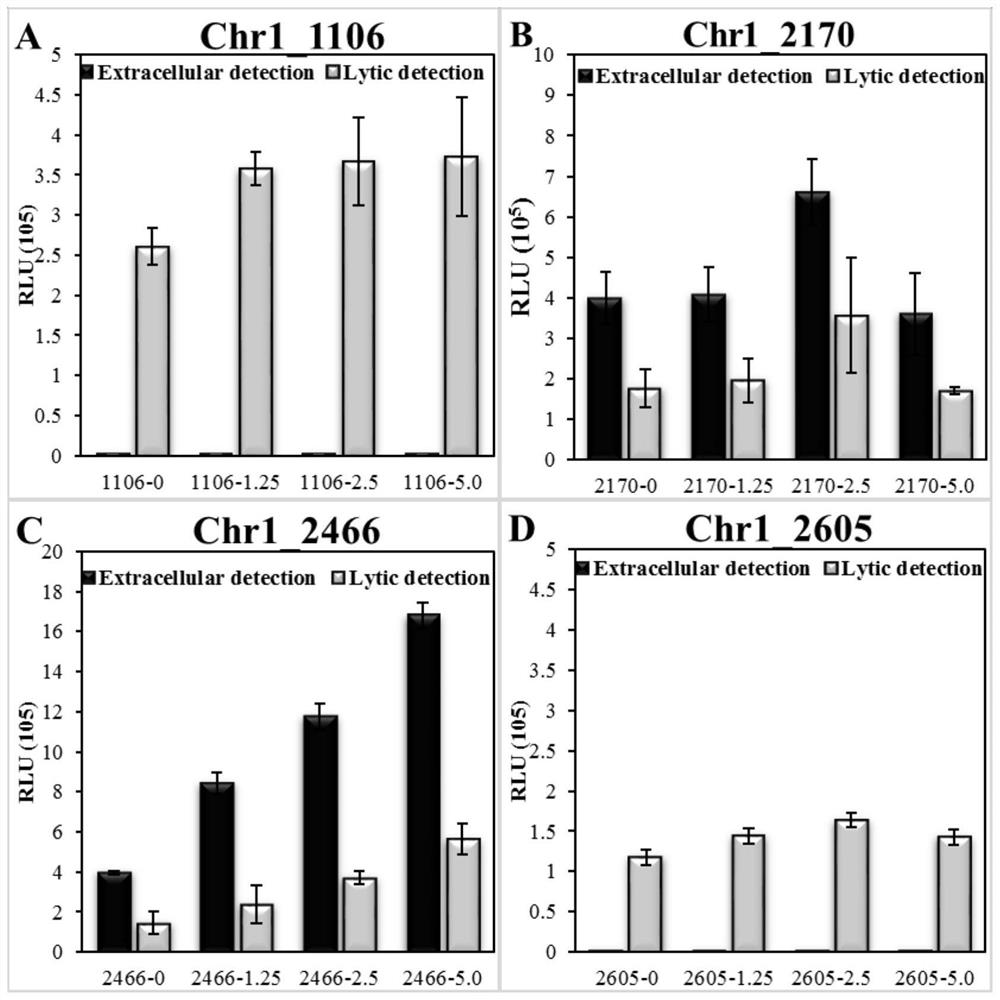
[0026] Example 2: Analysis of the activity and characteristics of polybrominated diphenyl ether recognition elements
[0027] Activity analysis was performed on the four candidate PBDE recognition gene elements obtained above. By synthesizing genes at Sangon Bioengineering (Shanghai) Co., Ltd., the coding sequence of firefly luciferase small peptide HiBiT (5′-GTGAGCGGCTGGCGGCTGTTCAAGAAGATTAGC- 3'), synthesis includes about 500bp upstream of each candidate gene including the gene of the promoter sequence, and designs BamHI and XhoI restriction sites at both ends of the gene fragment respectively (the 5' and 3' ends of chr1_1106 synthetic gene contain EcoRI and XhoI enzymes respectively cutting point). After gene synthesis, use TaKaRa's BamHI and XhoI restriction endonucleases to treat chr1_2605, chr1_2170 and chr1_2466 synthetic gene fragments and pET24a expression vectors in K buffer at 37 °C, use TaKaRa's EcoRI and XhoI restriction endonucleases in H buffer The chr1_1106 sy...
Embodiment 3
[0031] Example 3: Construction and performance analysis of a whole-cell microbial sensor for monitoring polybrominated diphenyl ethers
[0032] Synthetic primer 2466U (5'- GGATCC GCGGCGAAGGCATCTATAT-3') and primer 2466D (5'-GCGTCTTCCATTTCGGGATAATAGCC-3'), amplify chr1_2466 gene (its nucleotide sequence is as shown in SEQ ID NO.1) and its upstream 507bp sequence (its nucleotide sequence is as shown in SEQ ID NO.1) shown in ID NO.2). Primer LucU (5′-CGGCTATTATCCCGAAATGGAAGACGCCAAAAACA-3′) and primer LucD (5′- CTCGAG TTACACGGCGATCTTTCC-3') to amplify the complete sequence of the firefly luciferase luc gene (NCBI Accession No. AB762768.1). Using 2466U (5'- GGATCC GCGGCGAAGGCATCTATAT-3′) and primer LucD (5′- CTCGAG TTACACGGCGATCTTTCC-3') performed PCR fusion on the amplified chr1_2466 and luc gene fragments to obtain the fusion fragment chr1_2466-luc containing BamHI and XhoI restriction sites at both ends respectively. Use TaKaRa's BamHI and XhoI restriction endonucleases t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com