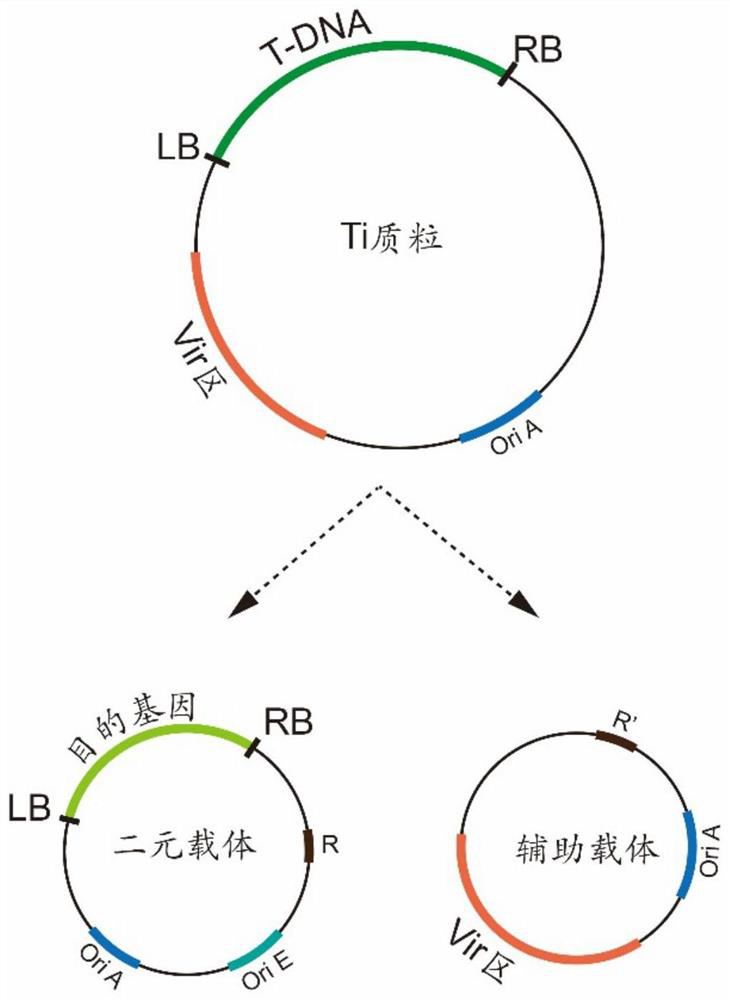
Method for screening single-copy T-DNA transgenic plant
A transgenic, single-copy technology, applied in the field of molecular biology, can solve the problems of amplification errors, lack of simple and reliable methods, and inability to distinguish two-fold differences in copy number, etc., achieving great application prospects, improving efficiency, and direct method. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The principle and design idea of embodiment 1SC PCR method
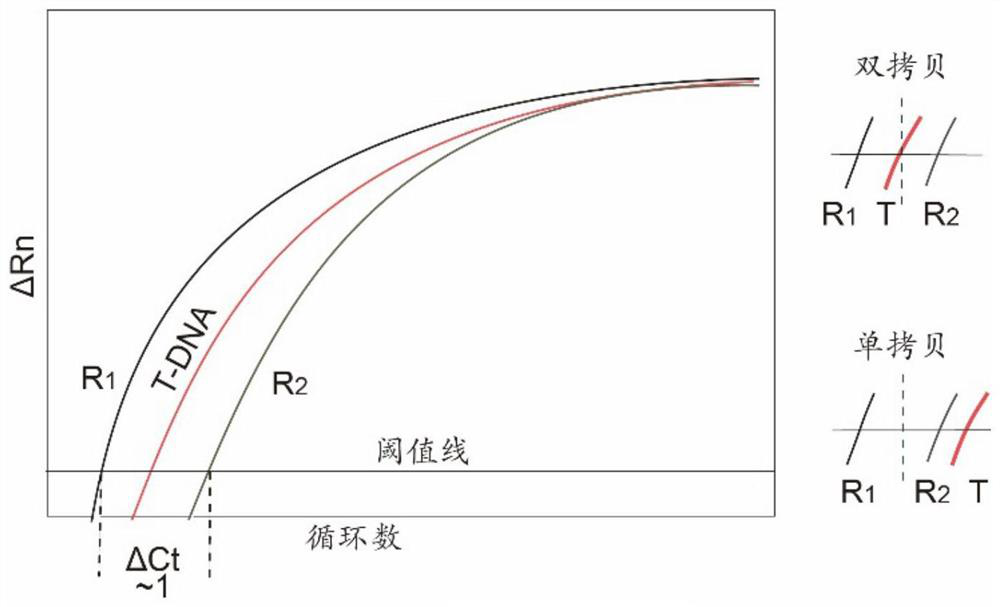
[0034] In the case of ensuring the amplification efficiency, the PCR amplification is close to 2 n Amplification was carried out (n is the number of cycles). Real-time quantitative PCR judges the copy number of the template by comparing the number of cycles (Ct value) at which the fluorescent signal of the amplified product reaches the threshold value. The number of templates amplified by a single-copy T-DNA template is reduced by half relative to that of a double-copy T-DNA, so the Ct value increases by 1. We use a pair of reference sequences (R 1 and R 2 ) to evaluate the change of Ct value of T-DNA. In our design, a pair of reference sequences R 1 and R 2 The amplification Ct values of the double-copy T-DNA plants are between R 1 and R 2 between, forming a sandwich of R 1 -T-R 2 The sandwich Ct relation of . Compared with double-copy plants, the Ct value of T-DNA amplification increases by ab...
Embodiment 2
[0035] Example 2 Establishment and verification of SC PCR method for screening single-copy T-DNA transgenic plants
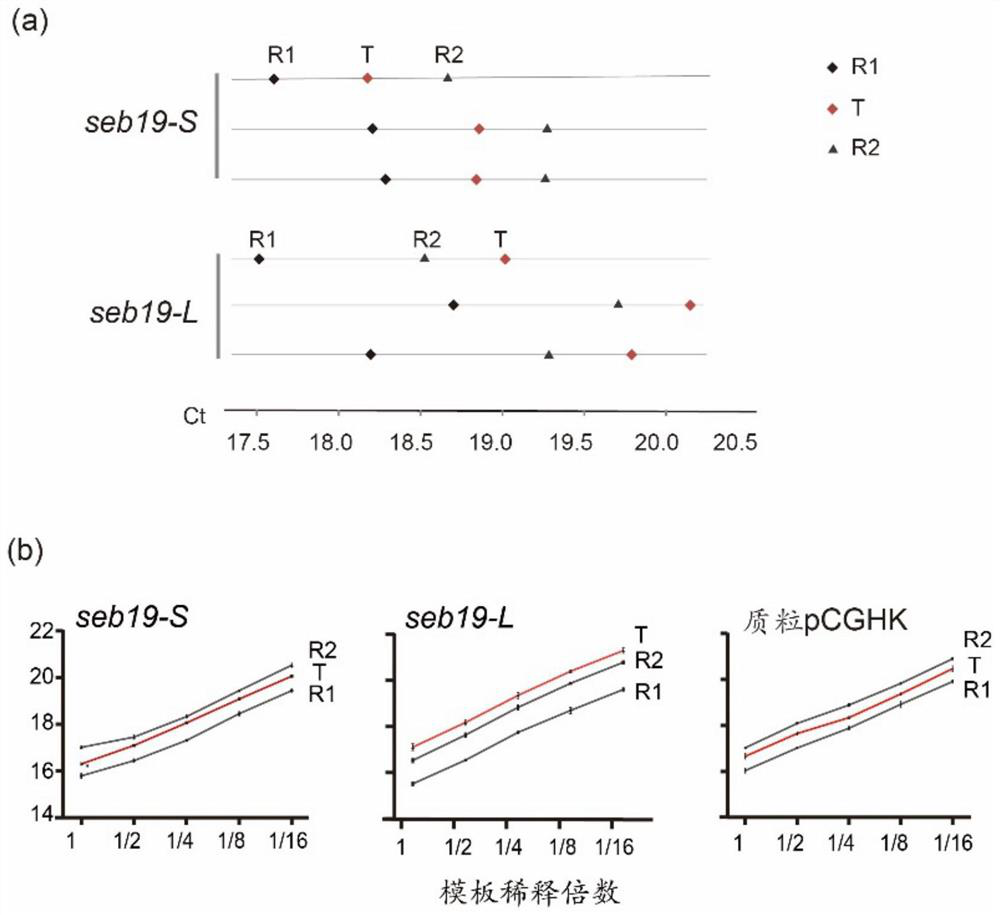
[0036] (1) We selected the transgenic plant seb19 inserted at a single site as the standard template (Hu,Y.,Chen, Z.,Zhuang,C.and Huang,J.(2017)Cascade of chromosome rearrangements caused by aheterogeneous T-DNA integration supports the double-stranded break repairmodel for T-DNA integration. Plant J, 90, 954-965.), to calibrate the sandwich relationship of Ct value. The seb19 transgenic plant has only one insertion site, and its progeny seb19-S is a homozygous plant with two T-DNA copies; while seb19-L is a heterozygous plant with only one T-DNA copy. The vegetative growth performances of the two offspring plants were different, and it was easy to distinguish them. The vegetative growth of seb19-L was consistent with wild type, while the vegetative growth of seb19-S was dwarf. We designed two pairs of reference sequences for the single-copy gene At1g06570 in ...
Embodiment 3
[0046] Embodiment 3 A method for obtaining single-copy T-DNA transgenic plants
[0047] (1) Obtaining Arabidopsis transgenic plants
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com