Application of MAL33 gene deletion in improving tolerance of saccharomyces cerevisiae to lignocellulose hydrolysate inhibitor
A technology of lignocellulose and Saccharomyces cerevisiae, applied in the direction of microorganism-based methods, applications, genetic engineering, etc., can solve the problem of low tolerance, achieve the effect of improving tolerance and shortening the lag period
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
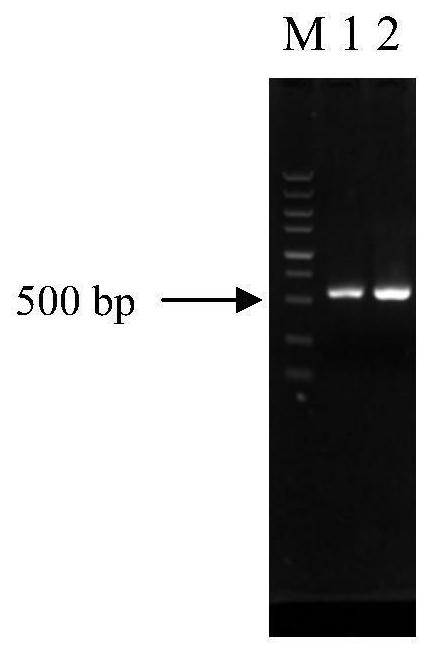
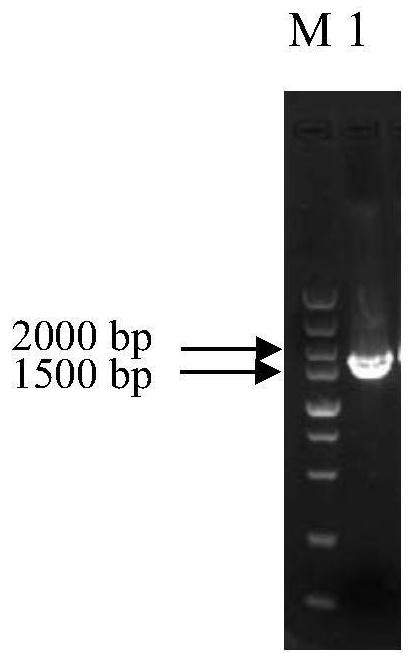
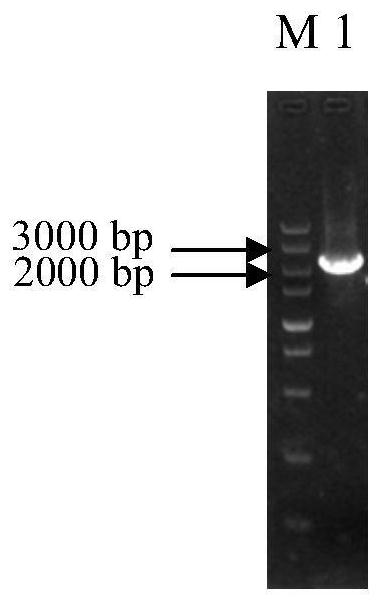
[0038] Example 1: MAL33 Construction of gene-deleted yeast strains
[0039] 1. Extraction of Saccharomyces cerevisiae genome
[0040] Saccharomyces cerevisiae BY4741 strain was cultured overnight in 5mL YPD medium, the cells were collected, and the BY4741 genomic DNA was extracted.
[0041] 2. Amplification of Gene Knockout Fragments
[0042] (1) MAL33 Amplification of the upstream homology arm and the downstream homology arm: using the genomic DNA of Saccharomyces cerevisiae BY4741 as a template, using primers UPS- MAL33 -F (5'-AATGGTCACTCCAAGTAACGGTATTGTGATTTCAACAGAA-3') and UPS- MAL33 -R (5'- TATTAAGGGTTGTCGACCTG ATCTTGACAACTGAGCTCTTTCACAC-3’) (underlined with loxP- KanMX4-loxP Homologous sequences upstream of the fragment for use with loxP-KanMX4-loxP for fusion PCR), and DOS- MAL33 -F (5'- TGATATCAGATCCACTAGTG TAGGACCCCTCATCACAATGATT -3’) (the underlined part is the loxP-KanMX4-loxP Downstream homologous sequences for use with loxP-KanMX4-loxP fusi...
Embodiment 2
[0048] Example 2: MAL33 Gene deletion strains are resistant to acetic acid, other typical inhibitors and H 2 o 2 tolerance test
[0049] 1. MAL33 Evaluation of the fermentation of gene-deleted strains in YPD medium containing 3 g / L acetic acid in oxygen-limited shake flasks
[0050] to pick MAL33 Gene deletion strain BSPX051-3XI- mal33△ and control strain BSPX051-3XI were inoculated into 5 mL of YPD liquid medium, activated and cultivated in a shaker at 30°C at 200 rpm for 12 h to 24 h, and then transferred to 5 mL of fresh medium after the bacterial liquid was cloudy Carry out the second activation, the activation time is 12 h~24 h. Inoculate the activated seed solution into an oxygen-limited bottle filled with 30 mL YPD + 3 g / L acetic acid medium, and adjust the initial OD 600 0.2, at 30 ℃, 200 rpm shaker in the oxygen-limited shake flask fermentation, sampling every few hours, using a UV-visible spectrophotometer to measure the OD of the fermentation broth 600 ,...
Embodiment 3
[0054] Example 3: MAL33 Oxygen-limited shake flask evaluation of gene-deleted strains in mixed sugar medium containing 3.5 g / L acetic acid and mixed sugar medium containing mixed inhibitors
[0055] 1. MAL33 Evaluation of the oxygen-limited shake flask fermentation of gene-deleted strains in YPDX medium containing 3.5 g / L acetic acid
[0056] to pick MAL33 Gene deletion strain BSPX051-3XI- mal33△ and control strain BSPX051-3XI were inoculated into 5 mL of YPD liquid medium, activated and cultivated in a shaker at 30 °C at 200 rpm for 12 h to 24 h, and then transferred to 5 mL of fresh medium after the bacterial liquid was cloudy Carry out the second activation, the activation time is 12h~24h. Inoculate the activated seed solution into 30 mL of YPDX medium containing 3.5 g / L acetic acid, adjust the initial OD 600 0.2, at 30 ℃, 200 rpm shaker in the oxygen-limited shake flask fermentation, sampling every few hours, using a UV-visible spectrophotometer to measure the OD ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com