Primer combination and application thereof in detection of lactobacillus paracasei in dairy products
A primer combination, dairy product technology, applied in the field of bioengineering, to achieve the effect of shortening the detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Example 1 A primer for detecting a via cheese lactoforma in a dairy product
[0053] (1) Primitive design principle
[0054] Using system development trees to homologously cluster analysis of amplification sequences, the GYRB gene amplification region of Lactobacillus CGMCC4691 is listed as a target, and system development trees are constructed; using ClustAlX1.81 to genbank lactic acid bacteria and its nearby The GyRB sequence is compared, and 30 sequences are selected;
[0055]After the above 30 sequences were compared with DAMBE 4.2.13 software, the same sequence was removed, and the system development trees were constructed using the 18 sequences retained;
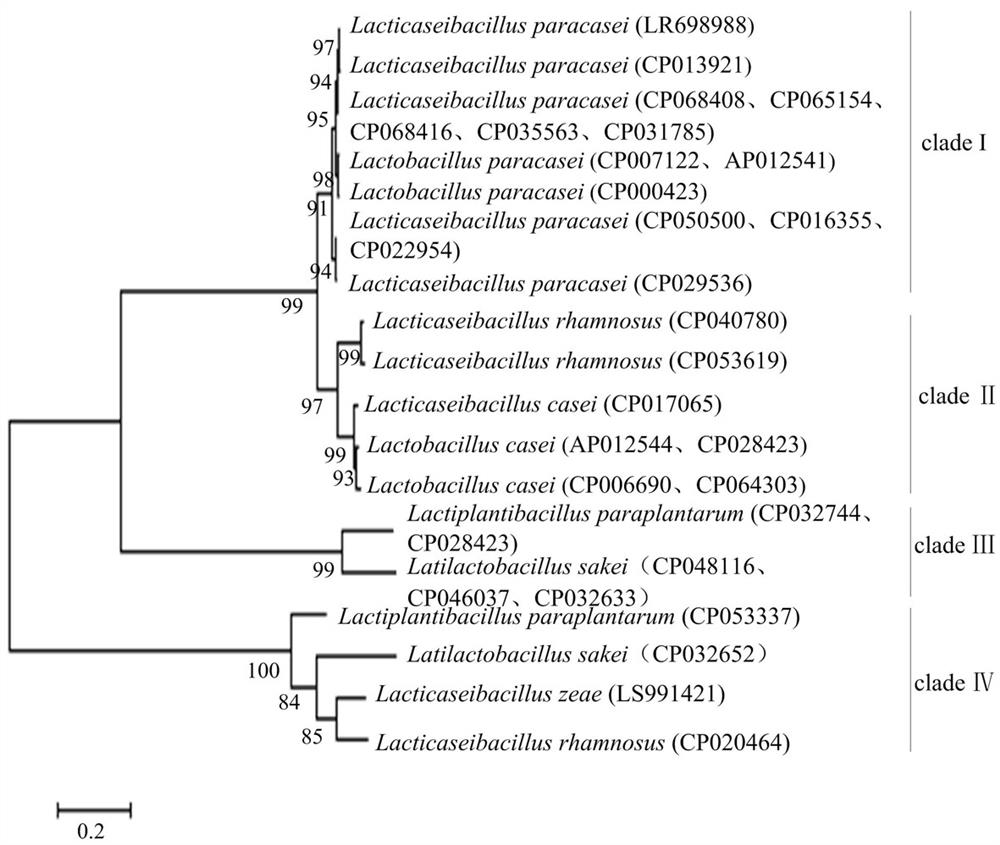
[0056] Based on the GYRB gene, the system development tree of L.Paracasei is built using the neighbor-joining method. figure 1 As shown, this tree is divided into four main branches: branch I, branch II, branch III and branch IV; branch i contains only a species, L.PaCTobacillus Paracasei; branch II consists of two ...
Embodiment 2
[0066] Example 2 Determination of auxiliary queen cheese emulsion heat
[0067] This example is a determination experiment of auxiliary cleaner heat tranout time, and the specific experimental step is:
[0068] A 1 mL of the sub-cheese lactofachalona was taken in 8 centrifugal tubes, and the centrifuge X1-X8 was removed, and the cured tube X1-X8 was treated with a water bath for 0min, 1 min, 2 min, 4 min, 6 min, 7 min, respectively. After 8 min or 9min, immediately cooled, and the number of sub-cheese lactobacillus in each dispersion was measured by a flat plate count method;
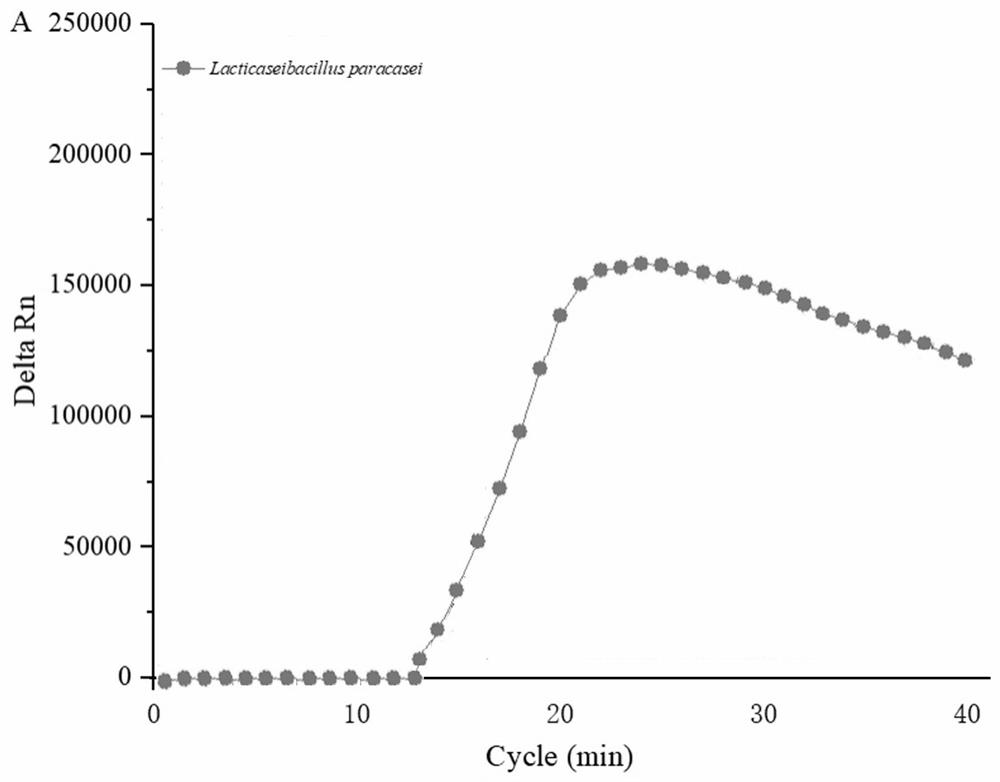
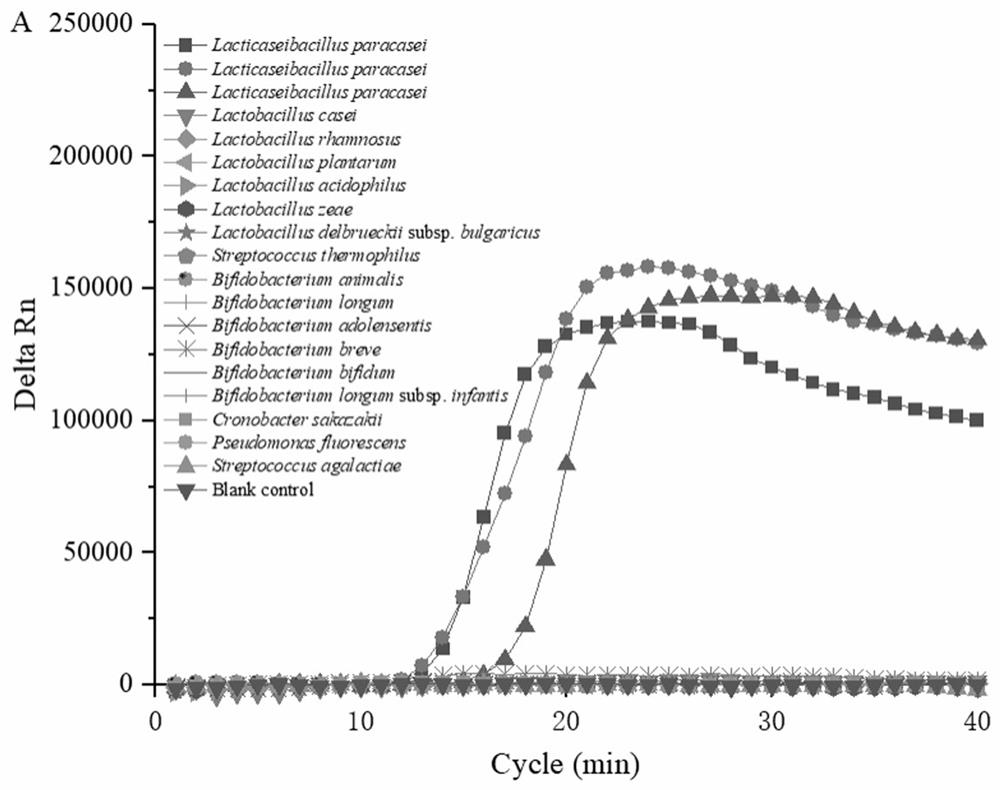
[0069] The results showed that the number of centrifugal tubes X1-X8 showed that the number of live bacteria was: 4.5 × 10, respectively. 8 CFU / ml, 2.3 × 10 6 CFU / ml, 1.2 × 10 6 CFU / ml, 4.8 × 10 3 CFU / ml, 1.7 × 10 2 CFU / mL, 6CFU / mL, 0CFU / mL, 0CFU / mL, which are known, heat treatment 8 min lactophilica all fire extinguishes, so the 90 ° C water bath is selected for 8 min to perform inactivatio...
Embodiment 3
[0070] Example 3 Optimization Experiment of PMA processing conditions
[0071] This example is an optimized experiment of PMA processing conditions, in order to obtain optimum PMA concentration, exposure time exposure time to the exposure time to eliminate the DNA amplification of the dead strain, according to Table 1 Design three factors three levels of orthogonal test, According to the aseptic water was washed twice with the aseptic water after treatment, the genomic DNA was subjected to 100 L of sterile water, and genomic DNA was extracted with a boiling water method, and the genomic DNA extracted by QLAMP was used, according to PMA pairs The inhibition rate of cheese DNA determines the best conditions for PMA treatment:
[0072] Table 1
[0073]
[0074] The specific experimental steps are:
[0075] The vicequarter CGMCC4691 was cultured in MRS medium at 37 ° C for 24 h, and the cells were rinsed with sterile saline, diluted to 10 5 CFU / ml, the bacterial suspension is name...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com