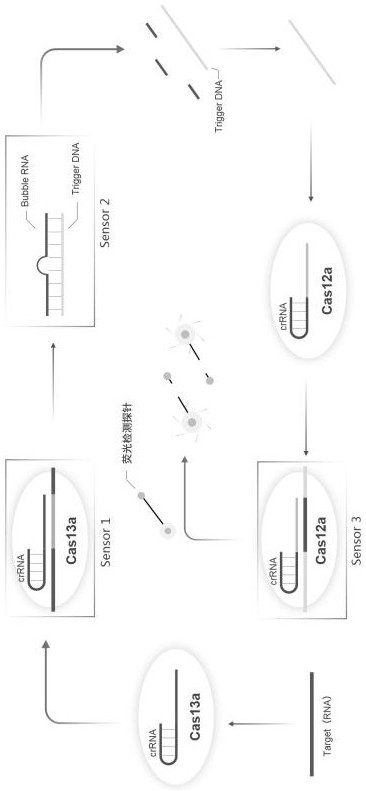
A kind of CRISPR cascade nucleic acid detection system and its detection method and application
A detection system and detection method technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, resistance to vector-borne diseases, etc., can solve the problems of sample loss, amplification deviation, false positive risk, etc., and achieve easy operation , short detection time and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] 1. Selection of target genome
[0081] Use NCBI to find the target sequence, use known information such as gene name or gene ID, and associate it with the corresponding Reference Sequence (RefSeq), so as to obtain the target sequence information, and finally use NCBI Blast to perform homology comparison to find the conserved sequence .
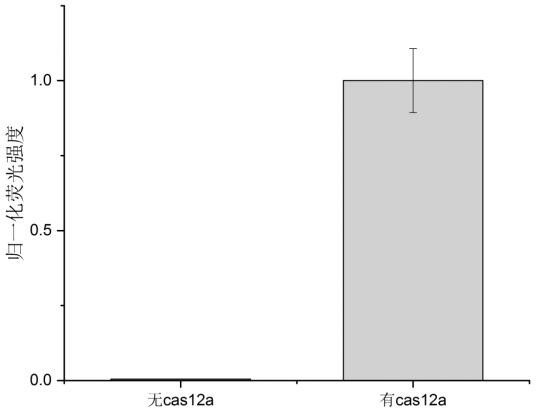
[0082] 2. Activity verification of Cas12 enzyme
[0083] Take the Lbcas12a protein as an example:
[0084] Activity verification of LbCas12a protein: design and synthesize a ssDNA probe labeled with a fluorescent group and a quencher group (specifically 5'-FAM-TTTTTT-BHQ1-3'), and prepare a reaction system based on the CRISPR-Cas system, For real-time fluorescence verification of trans-cleavage of LbCas12a.
[0085] The specific reaction system includes (30μL system): 60nM Lbcas12a, 60nM target guide 12-crRNA, 3× Cas buffer, 3μM ssDNA probe, 40nM target sequence, 1U / μL Murine RNase Inhibitor.
[0086] React at a constant temperature...
Embodiment 2
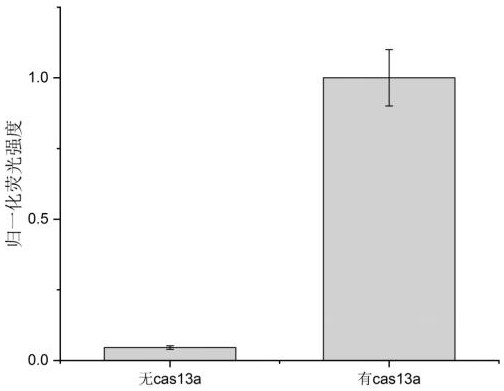
[0101] In order to further verify the feasibility of the nucleic acid detection method described in the embodiments of the present invention, a novel coronavirus (SARS-CoV-2) was selected for nucleic acid detection. It should be noted that the examples are for illustration only, and do not constitute any limitation to the protection scope of the present invention.
[0102] 1. Design the target guide RNA (13-crRNA) targeting the SARS-CoV-2 genome
[0103] The 13-crRNA targets the forward sequence of the SARS-CoV-2 genome, which is derived from the E genome of SARS-CoV-2. The forward sequence of 13-crRNA is: 5'-GAGACCACCCCAAAAAUGAAGGGGACUAAAAACCAAGACUCACGUUAACAAUAUUGCAGCA-3'.
[0104] The nucleic acid of the sample to be tested is synthesized by a third-party synthesis company. The selected fragment is the E gene fragment of SARS-CoV-2 (NCBI Reference Sequence: NC_045512.2), and its forward sequence is: 5'-UGCUGCAAUAUUGUUAACGUGAGUCUUG-3'.
[0105] 2. Prepare detection reaction...
Embodiment 3
[0112] In order to further evaluate the detection sensitivity of the nucleic acid detection method described in the embodiments of the present invention, the E gene fragment of SARS-CoV-2 is used as a target, and a series of serial dilutions (1 amol / L, 10 amol / L, 10 fmol / L, 10 pmol / L, 10nmol / L; Note: amol / L is 10 -18 mol / L), with no target SARS-CoV-2 genome as a negative control, nucleic acid detection was performed. The detection method and reagents are the same as in Example 2.
[0113] The result is as Figure 6 As shown, it can be seen that the CRISPR cascade nucleic acid detection method provided by the embodiment of the present invention can realize the detection of the E gene of SARS-Cov-2 as low as 1aM.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com