A DNA detection method based on CRISPR/Cas9 and its application
A detection method, DNA molecular technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., to achieve the effect of rapid DNA detection, wide application value, and avoiding nucleic acid hybridization and amplification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
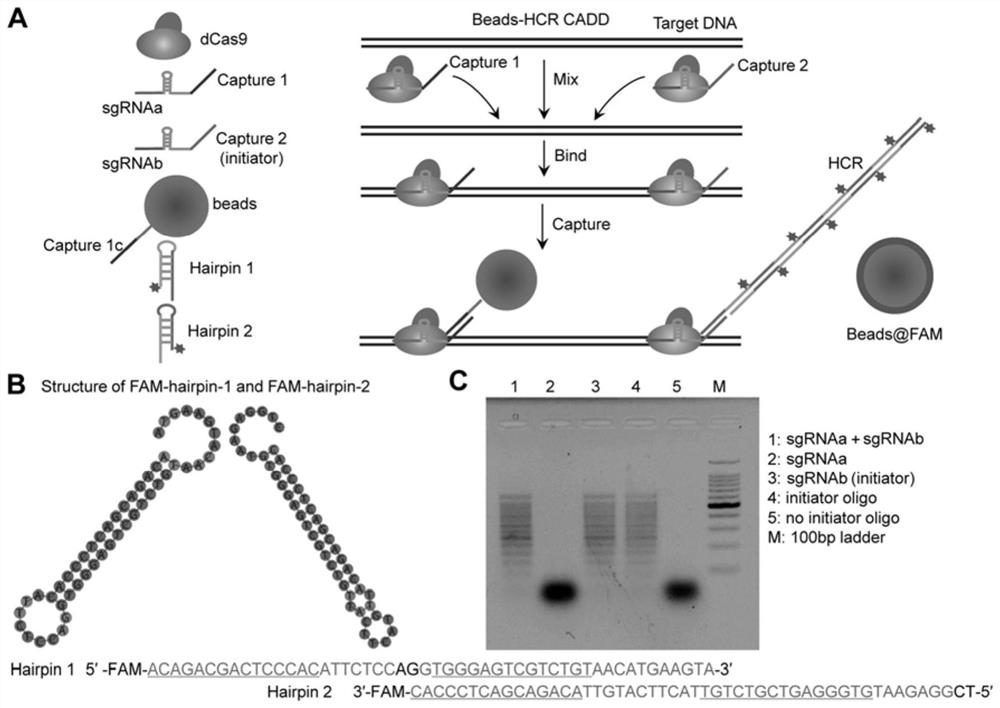
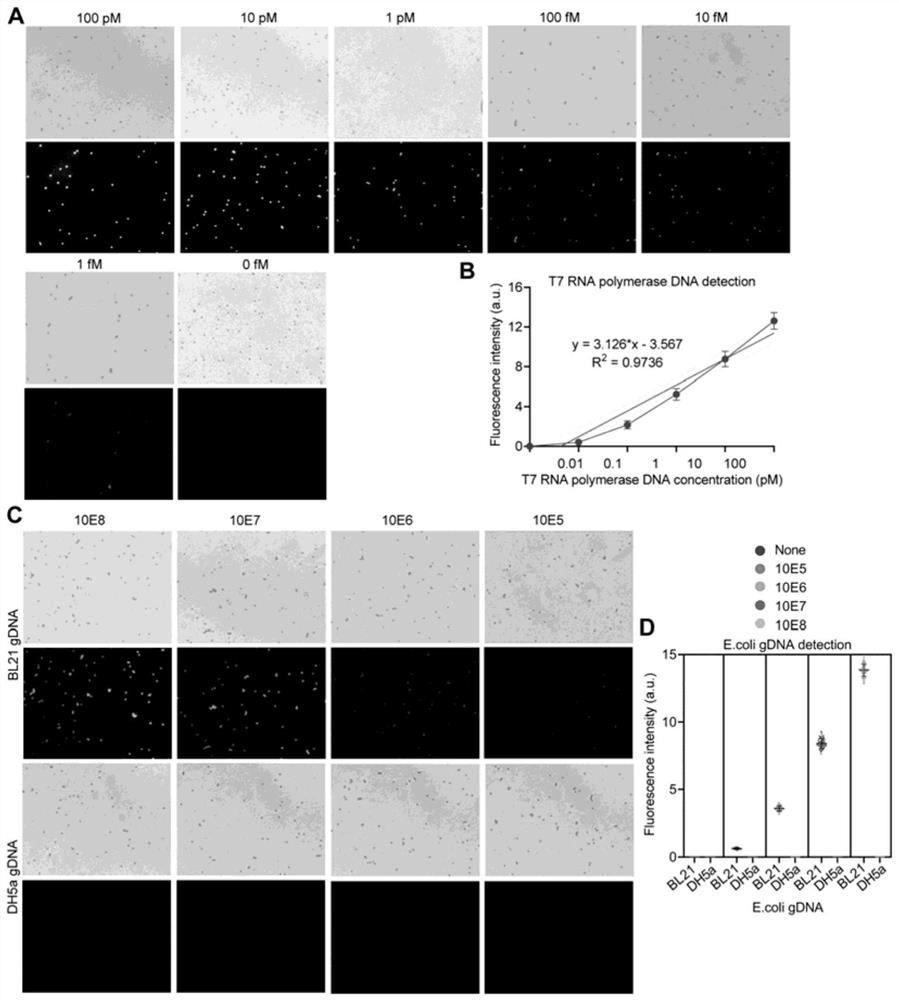
[0055] Detection of Escherichia coli T7 RNA polymerase with Beads-HCR CADD
[0056] 1.1. Experimental method:
[0057] 1.1.1. Preparation of sgRNA in vitro transcription template by PCR amplification method
[0058] Using the online sgRNA design software Chop-Chop ( http: / / chopchop.cbu.uib.no / ), using hg19 as the reference genome, design a pair of sgRNA for each DNA molecule to be tested, named sgRNAa and sgRNAb respectively, where sgRNAa is used to bind to the capture oligonucleotide (Capture 1c) immobilized on the surface of magnetic beads or microwell plates , sgRNAb is used to bind to the oligonucleotide that captures the reporter ( figure 1 A. Figure 10 A. Figure 12 A). According to the design results, PCR primers were synthesized for PCR amplification to prepare DNA templates for preparing sgRNA by in vitro transcription. In Example 1, a pair of sgRNAs (Table 1.1) were designed for the T7 RNA polymerase DNA to be tested, and then corresponding PCR primers (Tabl...
Embodiment 2
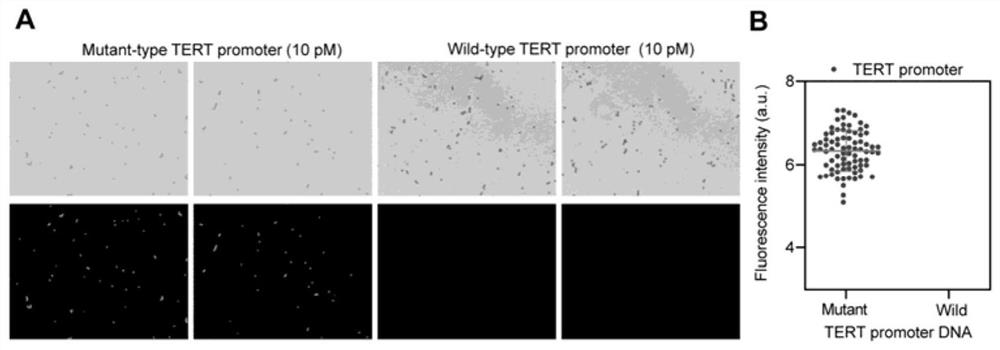
[0087] Detection of Mutant TERT Promoter DNA by Beads-HCR CADD
[0088] 2.1. Experimental method:
[0089] 2.1.1. Preparation of sgRNA in vitro transcription template by PCR amplification method
[0090] 1.1.1 with embodiment 1. The designed sgRNAs targeting mutant TERT promoter DNA are shown in Table 2.1. The primers for preparing sgRNA transcription templates by PCR are shown in Table 2.2.
[0091] Table 2.1, sgRNA targeting TERT promoter DNA (SEQ ID NO.36-37 in sequence)
[0092]
[0093] Table 2.2. PCR primers for preparing TERT promoter sgRNA in vitro transcription template
[0094] Primer name sequence (5'–3') F1, Ra, Rb, sgRa, sgRb Same as table 1.1 TERT-a-sgRNA-F2 GGACCGCGCTCCCCACGTGGGTTTTTAGAGCTAGAAATAGCAAG TERT-a-sgRNA-F3 TTCTAATACGACTCACTATAGGGACCGCGCTCCCCACGTGG TERT-b-sgRNA-F2 TCCCCGGCCCAGCCCCTTCCGTTTTAGAGCTAGAAATAGCAAG TERT-b-sgRNA-F3 TTCTAATACGACTCACTATAGTCCCCGGCCCAGCCCCTTCC
[0095] 2.1.2. Preparation of...
Embodiment 3
[0111] Detection of HPV DNA with Beads-HCR CADD
[0112] 3.1. Experimental method:
[0113] 3.1.1. Preparation of sgRNA in vitro transcription template by PCR amplification method
[0114] 1.1.1 with embodiment 1. The designed sgRNAs targeting 15 high-risk HPV (high-risk HPV, hrHPV) DNA are shown in Table 3.1. The primers for preparing sgRNA transcription templates by PCR are shown in Table 3.2.
[0115] Table 3.1. sgRNA targeting 15 kinds of hrHPV DNA. Identical list 1.
[0116] Table 3.2, PCR primers for preparing 15 kinds of hrHPV sgRNA in vitro transcription templates
[0117]
[0118]
[0119] 3.1.2. Preparation of sgRNA by in vitro transcription method
[0120] The preparation method is the same as 1.1.2 of Example 1.
[0121] The sgRNA prepared in Example 3 is used to detect HPV DNA, wherein the 5' end target DNA binding sequence of sgRNAa and sgRNAb is the same as the sequence SEQ ID NO.4-33 described in Table 1; the 3' end capture sequence of sgRNAa and s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com