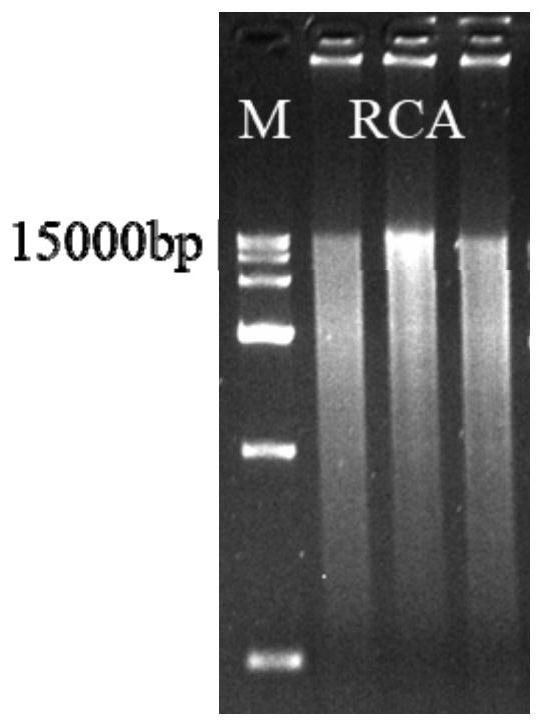
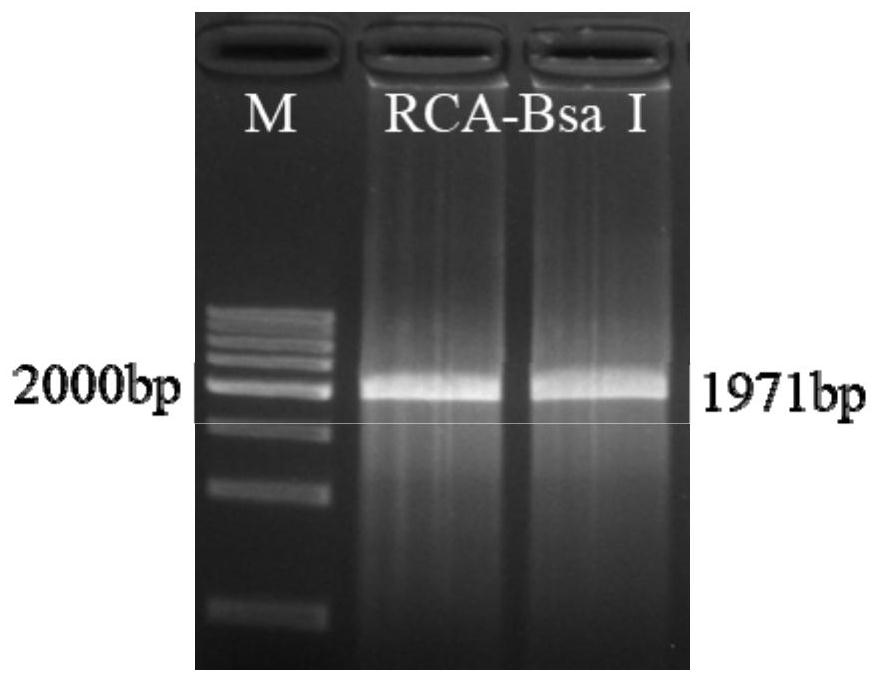
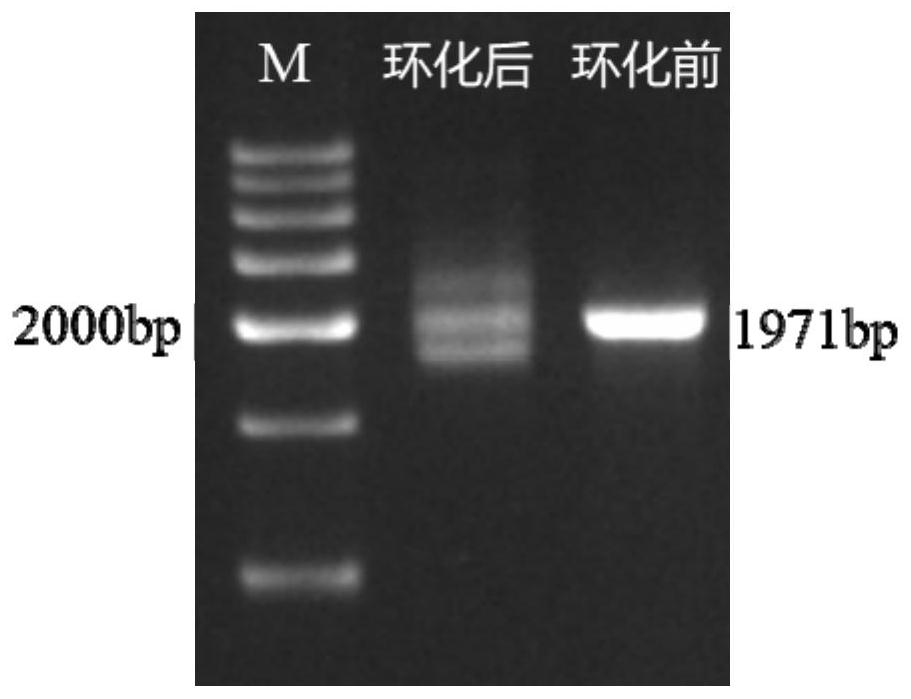
Method for preparing circular DNA in vitro
A circular and purposeful technology, applied in the field of in vitro preparation of circular DNA, can solve the problems of residual pollution process, complexity, high cost, etc., and achieve the effect of strong operability, simple process, and reduced biosafety risks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] 1. Select the green fluorescent protein (GFP) gene and its promoter (CMV) as the target linear DNA fragment.
[0033] 2. According to the CMV-GFP sequence information, design the Gibson assembly junction fragment according to "target DNA upstream 25bp + BsaI recognition site sequence + target DNA downstream 25bp". They are: cacat, ctgag, tacac, cgcct, acccg, acaca, cagaa, accgt, accca, tgccg, tttta, ccaga, caccc, gtggg, aggtg, ccaaa, attgg, ctgag, gacgg, gaata. The designed Gibson assembly connection fragments are as follows:
[0034]
[0035]
[0036]
[0037] 3. Synthesize the junction fragments by Gibson assembly, dilute to 10 μmol / L with sterile water, mix 10 μL each of the forward F and reverse R fragments, and anneal on the PCR instrument according to the following procedure to form double-stranded fragments: 95 ° C, 10 min; 85°C, 1min; 75°C, 1min; 65°C, 1min; 55°C, 1min; 45°C, 1min; 35°C, 1min; 25°C, 1min. After annealing, the 20 groups of fragments we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com