SNP (Single Nucleotide Polymorphism) for selecting piglets with high diarrhea resistance, detection method and application of SNP
A technology for piglets and piglet diarrhea, which is applied in the field of SNP selection of piglets with high anti-diarrhea ability, and can solve problems such as increasing the difficulty of vaccine development, drug residues, and enhanced bacterial resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Analysis of the promoter region of porcine STAT3 gene by PCR
[0050] 1. Genome extraction: The pig ear tissue was taken, and genomic DNA was extracted by conventional phenol-chloroform method.
[0051] 2. Primer design: Query pig STAT3 gene information (transcript ID: ENSSSCT00000018944.4) through NCBI's Ensemble database, download the 2500bp nucleotide sequence upstream of the first exon for primer design, the primers are STAT3F0 and STAT3R0, and the primer sequences are as follows :
[0052]
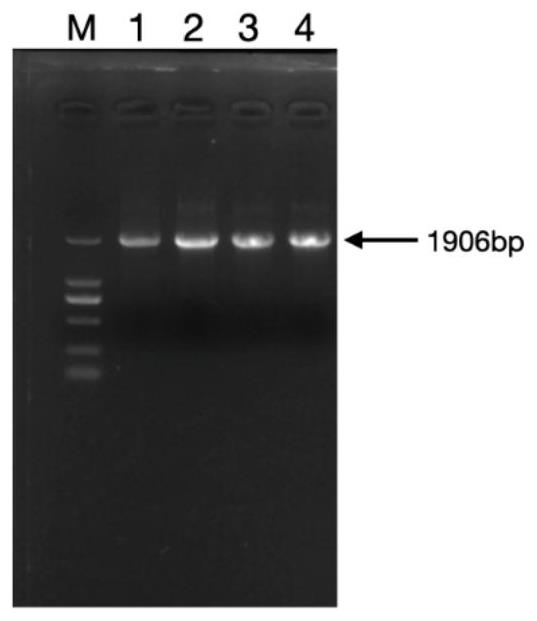
[0053] 3. PCR amplification: In this example, conventional PCR was used to amplify the -1134-+772 fragment of the promoter region of the STAT3 gene of Min pig. Amplification system: 100ng of genomic DNA, 25μL of 2×Taq Master Mix (Dye Plus), 10pmol of upstream and downstream primers, ddH 2 O make up to 50 μL. The amplification program was as follows: pre-denaturation at 94 °C for 5 min; denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s, extension at 72 °C for 2 m...
Embodiment 2
[0071] Msp I PCR-RFLP typing technique to detect the genotype of different breeds of pigs
[0072] 1. Establishment of genotyping technology:
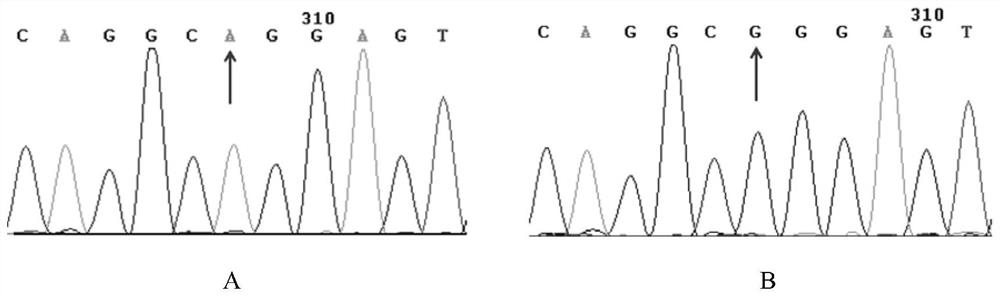
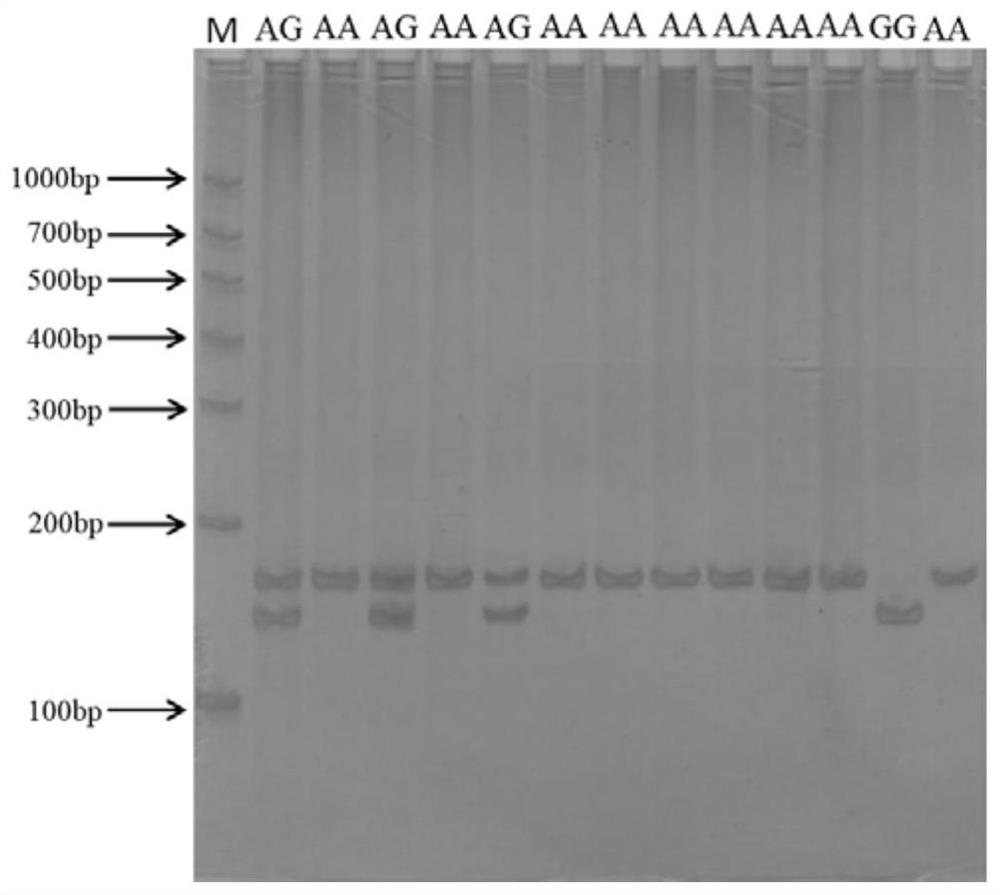
[0073] An A / G mutation was found at -870bp before the first exon of the STAT3 gene sequence. After the second base before the SNP site was mutated from G to C, Msp I (CCGG) forced enzyme could be generated cut point. After the PCR product was digested by Msp I enzyme, three band types appeared, respectively for the three genotypes: one band that was not recognized by Msp I enzyme was recorded as AA genotype (158bp), which could be recognized by Msp I enzyme. The identified two bands were GG genotype (137bp, 21bp), and the enzyme cut into three bands was recorded as AG genotype (158bp, 137bp, 21bp).
[0074] 2. Primer Design
[0075] According to the above analysis, STAT3 genotyping primers containing the SNP (NC_010454.4:rs327720240) site were designed. The primer sequences are as follows:
[0076]
[0077] 3. PCR amplification ...
Embodiment 3
[0086] Application of the molecular marker of the present invention in the correlation analysis of diarrhea in Min pig and Landrace pig suckling piglets
[0087] The experimental population used in this example came from Landrace pigs and Min pigs born in the same period in the autumn of 2013 in Lanxi County, Heilongjiang Province. From birth to 35-day-old weaning, check the anus of each suckling piglet at 6:00 am and 2:00 pm every day to observe whether there is fecal contamination, whether the anus is red and swollen, and the defecation of the piglets, and record and score. . The stool scoring standard is: the appearance of stool is strip or granular, the degree of diarrhea is normal, and the score is 0; the stool is soft and can be formed, and the degree of diarrhea is mild, and the score is 1; There was no separation of feces and water, the degree of diarrhea was moderate, and the score was 2; the feces were liquid, shapeless, and the feces and water were separated, and t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com