Bacteria screening reagent, screening method, kit and application
A screening method and kit technology, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., can solve the problems of inability to realize automatic replacement, increase throughput, disadvantages, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0067] Embodiment 1 lysozyme breaks bacteria
[0068] Raw materials: Lysozyme is purchased from Yisheng (egg white extraction, crystal powder), and the enzyme activity is (20KU / mg);
[0069] Reagent pre-mixed: 20mM Tris-HCl (pH 7.5) 500mL, 1mg mL -1 Lysozyme Tris-HCl (pH 7.5) solution (10×), stored in refrigerator at 4°C, 10mM EDTA;
[0070] 1) Prepare 1× lysis reaction lysozyme solution: take 1mL 1mg mL -1 After mixing lysozyme Tris-HCl (pH 7.5) solution with 9mL 20mM Tris-HCl (pH 7.5), add 0.1mL 10mM EDTA to make a mixed solution;
[0071] 2) Take the bacterial solution and centrifuge to remove the supernatant, add 1× lysis reaction lysozyme solution 10 times the volume of the precipitate, and resuspend by pipetting;
[0072] 3) React at 37°C and 600rpm for 5 minutes;
[0073] 4) Obtain the mixed bacterial liquid after lysis.
[0074] Due to the release of a large amount of DNA from the broken bacteria, the reaction system is viscous, which is not conducive to the pipet...
Embodiment 2
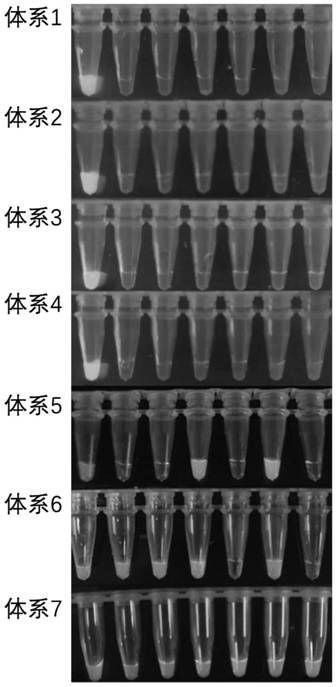
[0081] Embodiment 2 fluorescence detection
[0082] Table 2 Reaction system test result statistics
[0083] reaction system Test Results System 1 - System 2 - System 3 - System 4 - System 5 + / - System 6 + / - System 7 +
[0084] Table 3 System 1
[0085]
[0086]
[0087] Table 4 System 2
[0088] Reagent Dosage final reaction concentration 1 μM crRNA 1μL 50nM 10nM Cas12a protein 1μL 0.2nM 1nM fluorescent reporter 1μL 0.1nM 10*Buffer 2μL - ddH2O 13μL - Mixed bacteria 2μL -
[0089] Table 5 System 3
[0090] Reagent Dosage final reaction concentration 1 μM crRNA 2μL 100nM 10nM Cas12a protein 1μL 0.2nM 1nM fluorescent reporter 1μL 0.1nM 10*Buffer 2μL - ddH2O 12μL - Mixed bacteria 2μL -
[0091] Table 6 System 4
[0092] Reagent Dosage final reaction concentration ...
Embodiment 3
[0104] Embodiment 3 crRNA design
[0105] Different screening genes design different crRNAs. Select the PAM site (TTTN) on the target gene, select the 20bp base sequence after this site as the recognition sequence, and design crRNA.
[0106] 5′UAAUUUCUACUAAGUGUAGAU GAUUGCCGGACCCGGACCGC3 '(as shown in SEQ ID NO:1)
[0107] Wherein: the non-underlined part is the structural sequence, and the underlined part is the recognition sequence.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com