Ginkgo polymorphic EST-SSR (expressed sequence tag-simple sequence repeat) molecular marker primer and application thereof
A polymorphic primer and molecular marker technology, which is applied in the fields of bioinformatics, biochemical equipment and methods, and microbial determination/inspection, etc. Simple operation, wide coverage effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
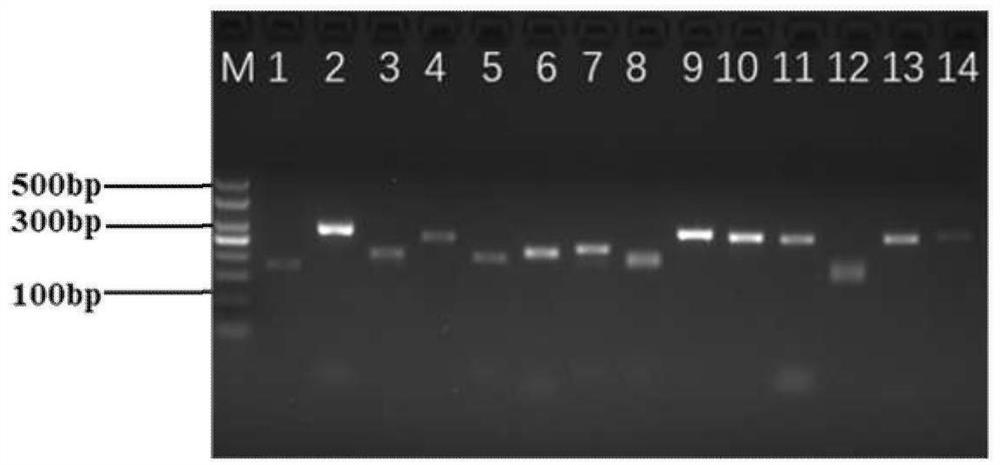
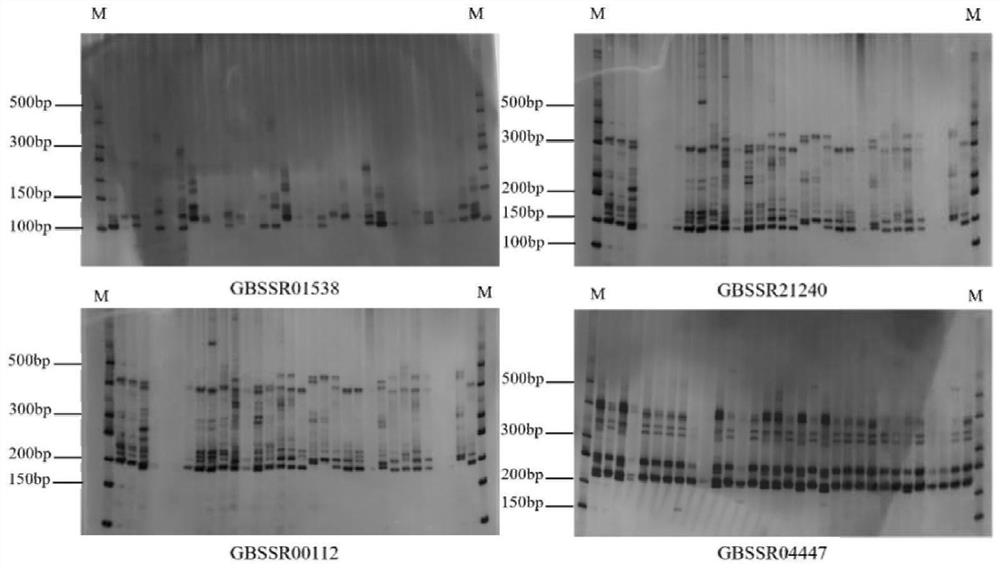
[0080] Embodiment 1: Development of Ginkgo biloba transcriptome EST-SSR molecular marker primers
[0081] 1 Materials and methods
[0082] 1.1 Materials
[0083]Based on the Ginkgo biloba transcriptome database of our group, the SSR loci were screened and analyzed. In the present invention, a total of 141 ginkgo germplasm resources are collected for the screening of polymorphic EST-SSR primers, of which 101 germplasm resources are known varieties (lines), and the remaining 40 are unknown varieties, according to "Huanglongxian No. 1-40" " to name it. 11 ginkgo varieties (strains) from Hunan were planted in the Ginkgo Experimental Demonstration Base of Xiling Industrial Zone, Damiaokou Forest Farm, Dong'an County, Hunan Province, and the remaining 90 varieties (series) of ginkgo were planted in the National Ginkgo Advanced Breed Base in Pizhou City, Jiangsu Province, with 40 copies. An unknown variety (strain) of ginkgo was planted in Huanglongxian Village, Jiangning District...
Embodiment 2
[0115] Embodiment 2: Genetic diversity analysis of ginkgo germplasm resources
[0116] 1 Materials and methods
[0117] 1.1 Materials
[0118] The test materials were the same as those in Example 1, and were 141 ginkgo germplasm resources.
[0119] 1.2 Methods
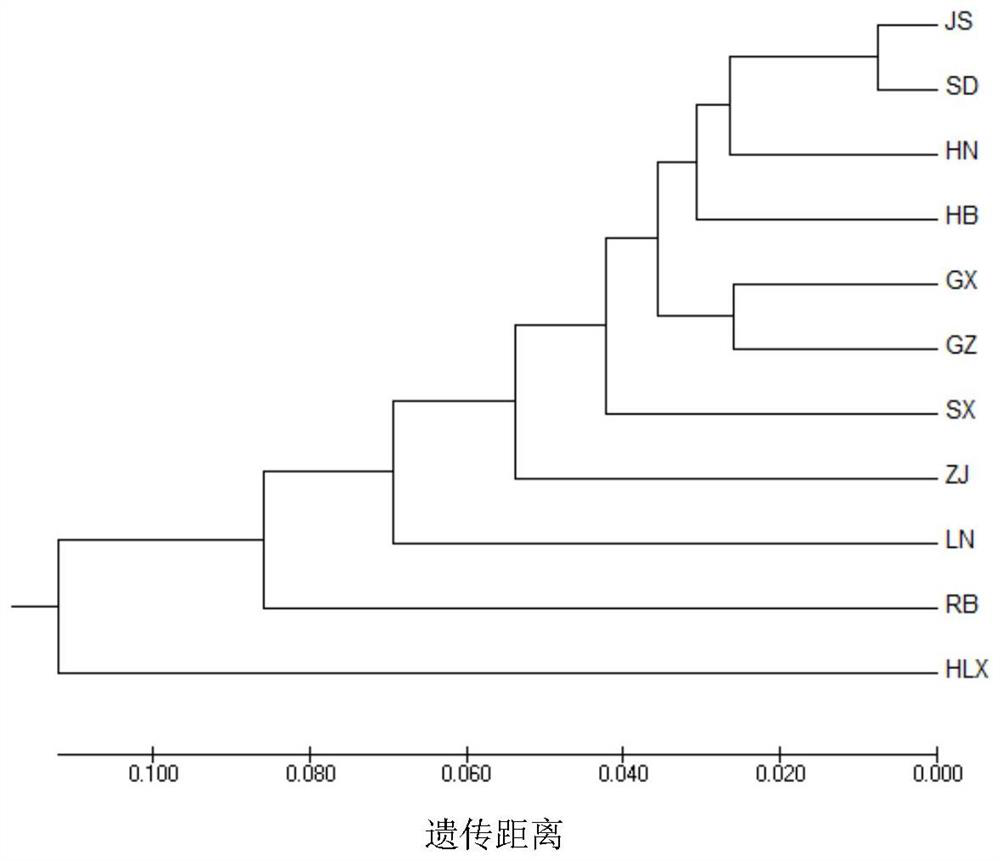
[0120] The 20 pairs of polymorphic EST-SSR molecular marker data obtained above were input into Popgene 32 software, and the average expected heterozygosity or gene diversity index (H) was calculated. e ), Shannon diversity index (I), genetic differentiation coefficient (G st ) and gene flow (N m ). Molecular variance analysis was performed by GenAlE_v6.502 software, and UPGMA cluster analysis was performed by NTSYS software.
[0121] 2 Test results
[0122] 2.1 Genetic diversity analysis of ginkgo germplasm resources
[0123] The results of genetic diversity analysis showed that (Table 5), 141 Ginkgo germplasm resources had a high level of genetic diversity. Gene Diversity Index (H e ) was between 0.328 and ...
Embodiment 3
[0138] Example 3: Construction of Ginkgo Core Germplasm
[0139] 1 Materials and methods
[0140] 1.1 Materials
[0141] The test materials were the same as those in Example 2, and were 141 ginkgo germplasm resources.
[0142] 1.2 Methods
[0143] 1.2.1 Construction of core germplasm
[0144] In order to compare the differences between different sampling methods and obtain the best sampling strategy, two grouping principles (grouping by origin, completely random without grouping), four in-group sampling ratios (simple ratio, logarithmic ratio, square root ratio and genetic diversity) were analyzed. sex ratio), 2 sampling methods (site-priority cluster sampling, random cluster sampling), 6 overall sampling ratios (5%, 10%, 15%, 20%, 25%, 30%, 35% , 40%) in any combination to form a total of 72 sampling schemes. The genetic distance is calculated based on the SM similarity coefficient.
[0145] 1.2.2 Evaluation of core accessions
[0146] By the number of alleles, the num...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Total length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com