High-efficient expression D-amino acid oxidase methanol yeast, its construction and fermentation method
A methanol yeast, high-efficiency expression technology, applied in the field of bioengineering, can solve problems such as low yield and difficult post-processing, and achieve the effects of low cost and simple fermentation conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
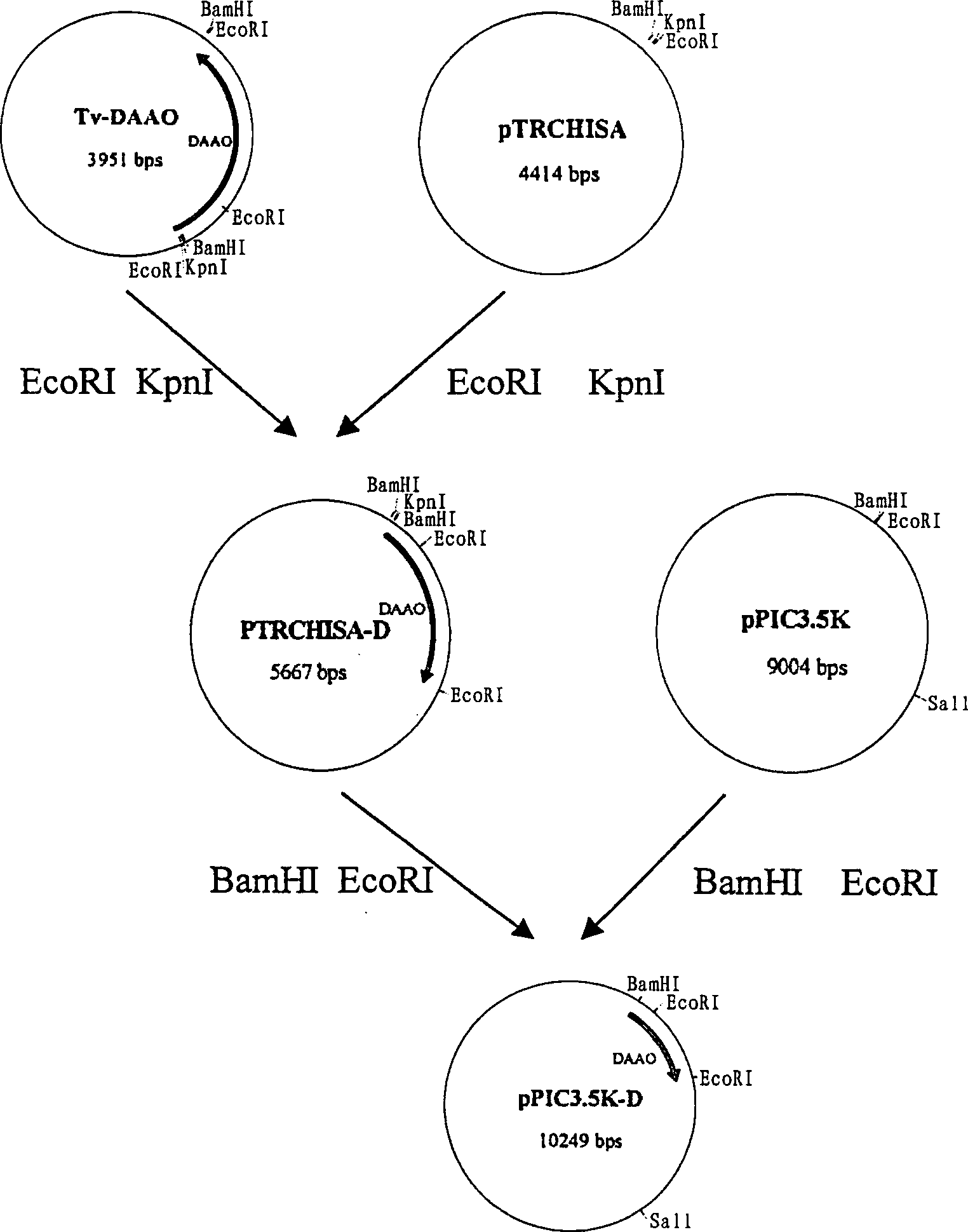
[0017] Example 1 Acquisition of D-amino acid oxidase gene
[0018] According to the known 5' and 3' end sequences of Trichomonas oxidase gene, the primers were designed as follows:
[0019] 5'-primer (introduced BamH I restriction site):
[0020] 5'- GGATCC ATGGCTAAAATCGTTGT-3'
[0021] 3'-primer (introduced EcoR I restriction site):
[0022] 5'- GAATTC GTTGTTGATGGGAGGTAA-3'
[0023] Plasmid pAO6 containing the Trichomonas D-amino acid oxidase gene with the intron region removed was kindly provided by Prof. Dominguez. Using the plasmid pAO6 as a template, under the action of Taq DNA polymerase, the D-amino acid oxidase gene of Trichomonas spp. was transcribed and synthesized by PCR, and its 5' and 3' ends had BamHI and EcoR I restriction enzyme sites, respectively. The reaction conditions are:
[0024] 94°C, 30sec 5 cycles 94°C, 30sec 30 cycles 94°C 5min→→40°C, 30sec→→→→ 48°C, 30sec→→→→72°C, 10min
[0025] 72°C, 75sec 72°C, 75sec
[0026] After the P...
Embodiment 2
[0027] A total of 15ul of the above mixture was reacted overnight at 16°C. The resulting ligation product was transformed into E.coli TG1 competent cells, and spread on an ampicillin plate to screen positive clones. Plasmid DNA was extracted using a plasmid extraction kit (Promega product), and the obtained plasmid DNA was identified by PCR to prove that the recombinant plasmid was T-vector-DAAO.
Embodiment 3
[0028] Example 3 Construction of D-amino acid oxidase gene recombinant plasmid pTRCHISA-DAAO
[0029] Digest the T-vector-DAAO plasmid with BamH I, and cut out a DNA band with a size of about 1.3Kb, which proves that the D-amino acid oxidase gene is inserted in reverse. Digested with Kpn I, then incompletely digested with EcoR I, electrophoresed on 1% agarose, and a DNA gel recovery kit was used to recover a D-amino acid oxidase gene fragment of about 1.3Kb. The recovered fragment was ligated with the pTRCHISA plasmid cut with the same restriction enzymes, and the ligation reaction system was as follows:
[0030] pTRCHISA (Kpn I, EcoR I cut) 1ul
[0031]D-amino acid oxidase gene fragment 3ul
[0032] 5× ligation buffer 3ul
[0033] T4 DNA Ligase (1u / ul) 1.5ul
[0034] h 2 O 6.5ul
[0035] A total of 15ul of the above mixture was reacted overnight at 16°C. The resulting ligation product was transformed into E.coli TG1 competent cells, and spread on an ampicillin plate to...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com