BZIP type transcripton factors regulating expression of rice storage protein
A protein and plant transformation technology, applied in anti-plant immunoglobulin, hydrolase, genetic engineering, etc., can solve the problems of expression mode change, promoter activity reduction, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0111] [Example 1] Isolation of a cDNA clone encoding a bZIP transcription factor from a seed cDNA library
[0112] The leaves and roots of rice plants (Oryza sativa L.c.v. Mangetumochi) cultured in hydroponic culture for 14 days were frozen in liquid nitrogen and stored at -80°C until use. Mature rice seeds were collected from rice plants grown in the field.
[0113] Oligonucleotide primers were designed according to the highly conserved amino acid sequence (SNRESA and KVKMAED) in the bZIP region of Opaque2(O2)-like protein, and poly(A) extracted from rice seeds + mRNA was used as template for RT-PCR. Poly A RNA was extracted from seeds 6 to 16 days after anthesis (DAF) (Takaiwa F. et al., Mol. Gen. Genet. 208:15-22, 1987), using Superscript reverse transcriptase (Gibco BRL, Paisly, UK) synthesized single-stranded cDNA by reverse transcription, and oligo(dT) 20 for primers. The cDNA was then amplified using the following primer pairs: 5'-TCC AAC / TA / CGI GAA / GA / TCIGC-2'; SE...
Embodiment 2
[0115] [Example 2] Identification of RISBZ cDNA
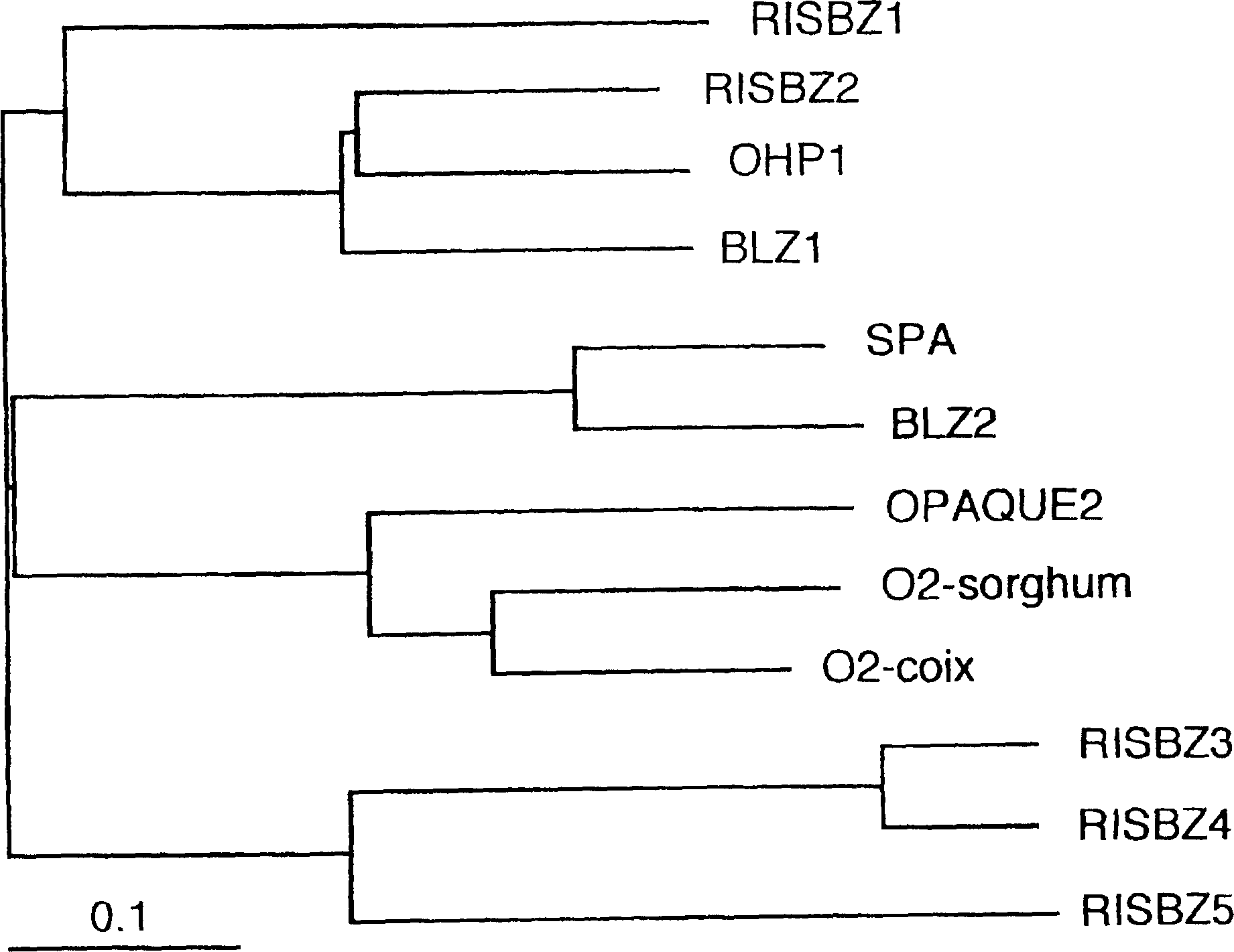
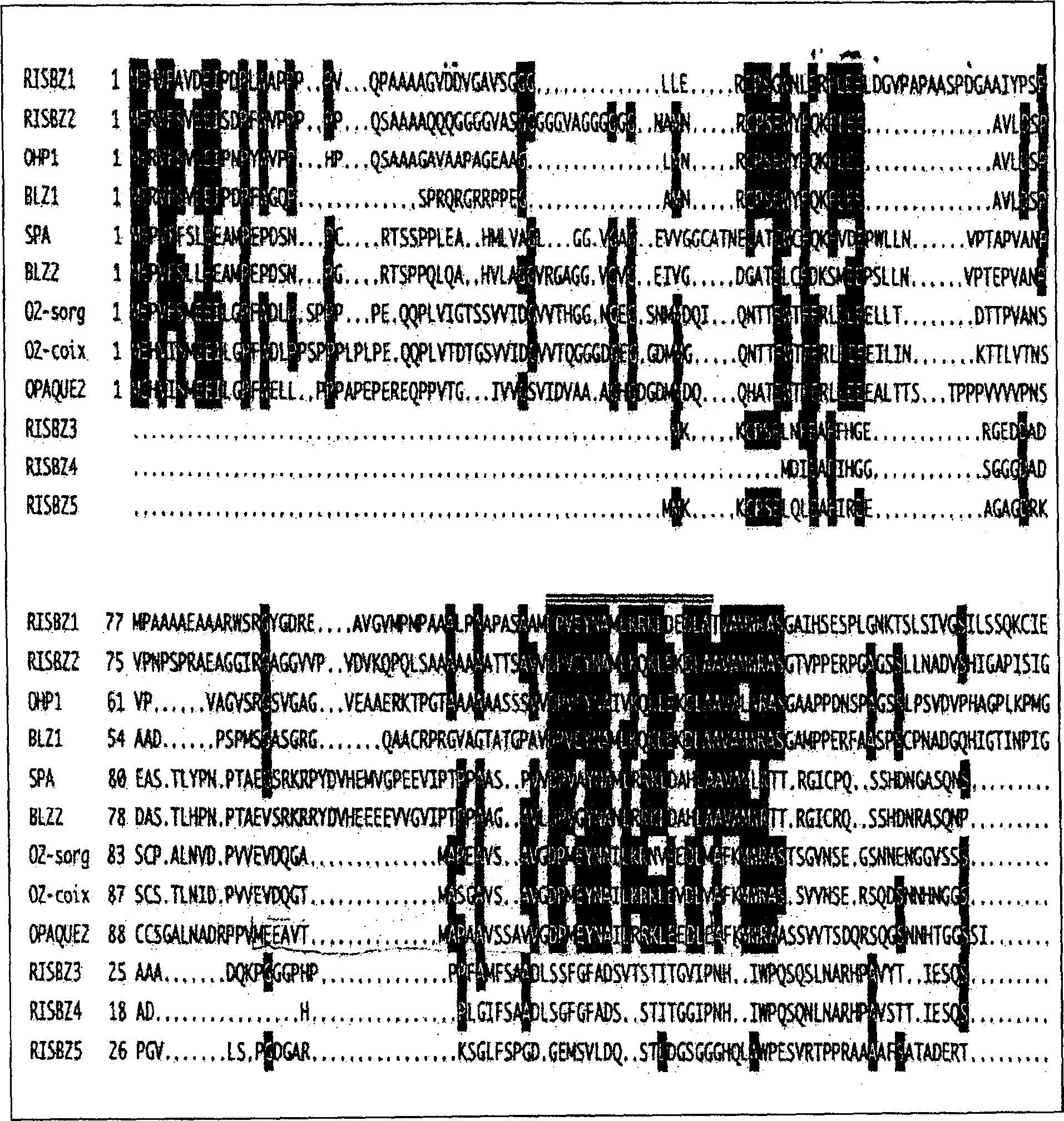
[0116] The newly confirmed RISBZ cDNAs (RISBZ1, RISBZ4, RISBZ5) were identified in detail as follows. RISBZ1 cDNA is the longest, 1742bp excluding polyadenylic acid, and contains a reading frame encoding 436 amino acids (estimated molecular weight is 46,491Dal). RISBZ4 and RISBZ5 have reading frames encoding 278 and 295 amino acids; their predicted molecular weights are 29383 and 31925 Dal, respectively.
[0117] RISBZ1 mRNA has a leader sequence (245 bases long) that is longer than the average leader sequence. Interestingly, a small open reading frame encoding 31 amino acid residues was found within the leader sequence upstream of the actual start codon of the RISBZ1 protein. Previous studies on maize Opaque2(O2) (Hartings H. et al., EMBO J.8: 2795-2801, 1989), wheat SPA (Albani D. et al., Plant Cell 9: 171-184, 1997), barley BLZ1 and BLZ2 (Vincente -Carbojos J. et al., Plant J.13: 629-640, 1998; Onate L. et al., J. Biol....
Embodiment 3
[0123] [Example 3] Genome structure of RISBZ1 gene
[0124] Using primers designed from the nucleotide sequence of RISBZ1 cDNA, the genomic region encoding the promoter and RISBZ1 protein was isolated. A PCR reaction was performed using rice genomic DNA as a template and two pairs of oligonucleotides as primers (RIS1f: 5'-ATGGGTTGCGTAGCCGTAGCT-3' / SEQ ID NO: 18 and RELr5: 5'-TTGCTTGGCATGAGCATCTGT-3' / SEQ ID NO: 19) and (RELf2: 5'-GAGGATCAGGCCCATAT-3' / SEQ ID NO: 20) and RIS1r: 5'-TCGCTATATTAAGGGAGACCA-3' / SEQ ID NO: 21). TAKARALA Taq polymerase (TAKARA) was used to amplify DNA fragments in a thermal cycler, using 30 cycle reactions, 98°C for 10 seconds, 56°C for 30 seconds, and 68°C for 5 minutes. The promoter region of the RISBZ1 gene was also amplified by thermal asymmetric staggered (TAIL) PCR based on the method of Liu et al., in which three oligonucleotides were used as specific primers, tail1: 5'-TGCTCCATTGCGCTCTCGGACGAG-3' / SEQ ID NO:22, tail2:5'-ATGAATTCGCGAGGGGTTTTCGA-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com