Combinatorial libraries of proteins having the scaffold structure of c-type lectin-like domains
A technology of lectin-like and scaffold structure is applied in the field of protein combinatorial library with scaffold structure of C-type lectin-like domain, which can solve the problem of increasing the number of CTLDs and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0413] Construction of Escherichia coli expression plasmids and phagemids derived from tetrapodrin
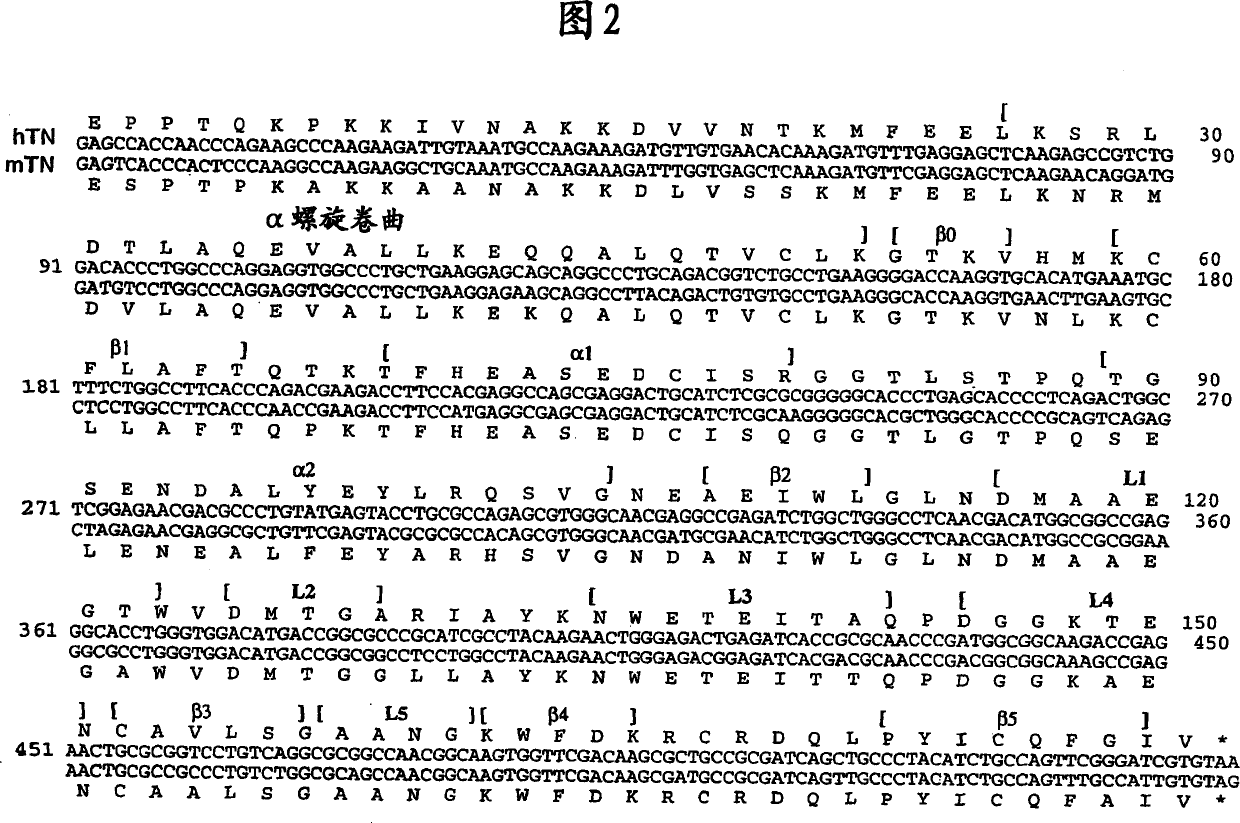
[0414] Take advantage of QuickChange TM Site-directed mutagenesis kit (Stratagene, La Jolla, CA) and performed as described in the manual, starting with the expression plasmid pT7H6-rTN 123 [Holtet et al. (1997)], 4 consecutive series of site-directed mutagenesis experiments were performed, An expression plasmid pT7H6FX-htlec encoding the FX-htlec (SEQ ID NO: 01 ) portion of the full-length H6FX-htlec fusion protein was constructed. Mismatched primer pairs to introduce the desired mutations were supplied by DNA Technology (Aarhus, Denmark). Figure 5 shows a schematic diagram of the obtained pT7H6FX-htlec expression plasmid, and SEQ ID NO: 01 gives the nucleotide sequence encoding the insert segment of FX-htlec. Figure 6 shows the amino acid sequence of the FX-htlec portion of the H6FX-htlec fusion protein and is given in SEQ ID NO:02.
[0415]By amplifying and subcloning int...
Embodiment 2
[0422] Confirmation of successful display of Phtlec and PhTN3 on phage
[0423]In order to verify that the fusion protein of Phtlec and PhTN3 Gene III can indeed be displayed using recombinant phage particles, the phagemids pPhtlec and pPhTN3 (described in Example 1) were transformed into Escherichia coli TG1 cells and infected with helper phage M13KO7 to produce Recombinant phages were obtained. Recombinant phages were separated by precipitation with polyethylene glycol (PEG8000), and ELISA-type "sandwich" tests were performed on Phtlec and PhTN3 phage preparations and helper phage samples. Protein and bovine serum albumin (BSA) and blocked in skim milk or skim milk / EDTA. Briefly, pPhtlec and pPhTN3 phagemid-transformed TG1 cell cultures were grown at 37°C in 2×TY-medium supplemented with 2% glucose and 100 mg / L ampicillin until their A 600 reach 0.5. At that time, helper phage M13KO7 was added to it to a concentration of 5 × 10 9 pfu / mL. Cultures were incubated for an a...
Embodiment 3
[0427] Confirmation of the authentic ligand-binding properties of Phtlec and PhTN3 displayed on phage
[0428] The apogroup of the CTLD domain of human tetramantin binds specifically to the human plasminogen kringle 4 domain in a lysine-sensitive manner [Graversen et al. (1998)]. Tetramantin binding to plasminogen is inhibited by calcium or lysine analogs such as AMCHA (6-amino-cyclohexanoic acid), where calcium binds to two of the ligand-binding sites of the CTLD domain point (Kd is about 0.2mM), while lysine analogs can specifically bind to two stronger lysine binding sites in plasminogen (Kd is about 15mM), these two sites One is positioned in kringle 1 and the other is positioned in kringle 4.
[0429] To demonstrate the specific AMCHA-sensitive binding of human Phtlec and PhTN3 phage to human plasminogen, an ELSISA assay similar to that applied to demonstrate the presence of Phlec and PhCTLD GIII fusion proteins displayed on phage particles (see Example 2) was designed. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com