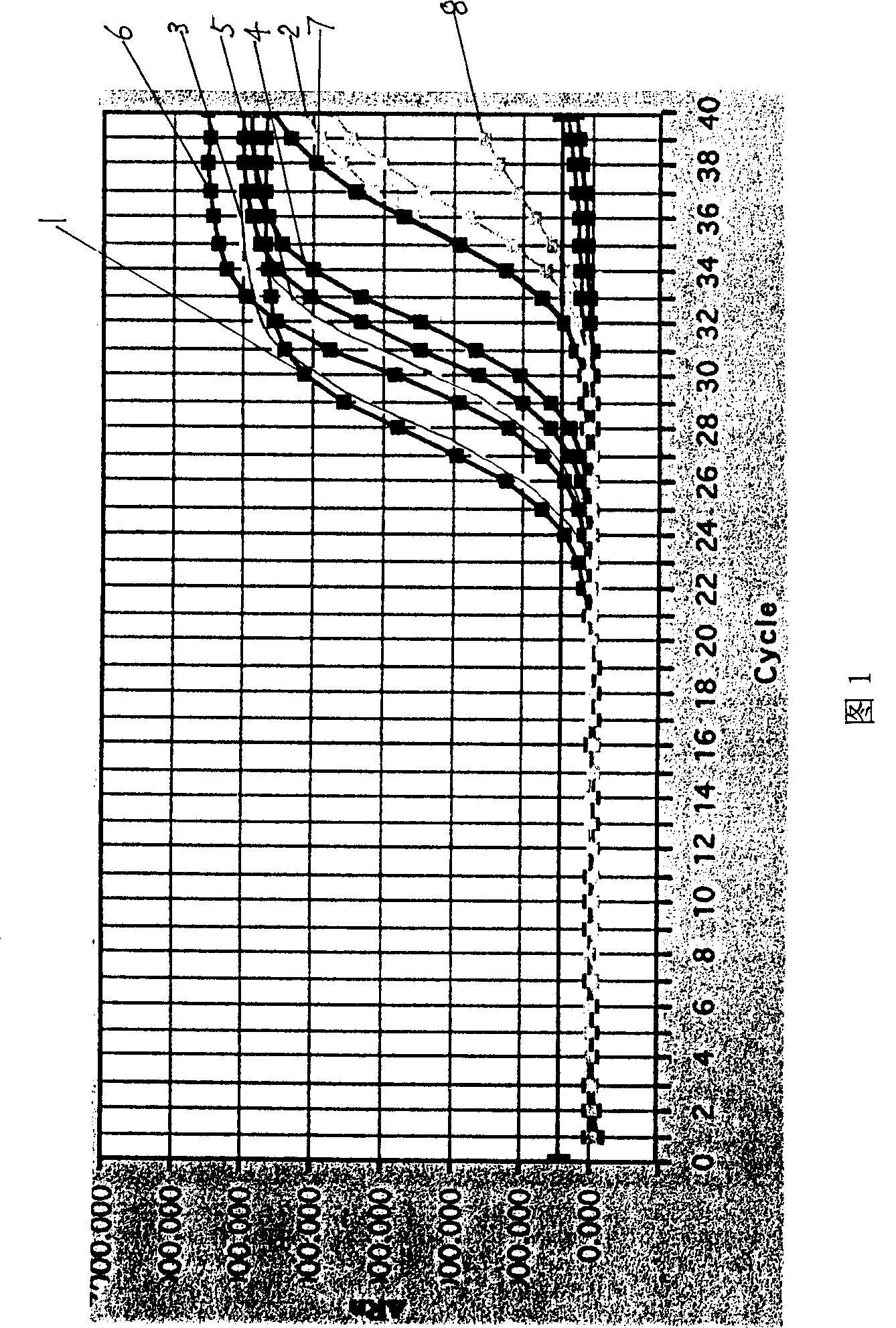
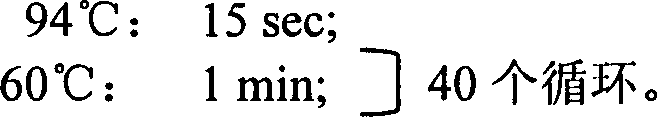Probe sequence for qualitatively detecting transgenic crop containing cauliflower mosaic virus 35S promotor using fluorescence PCR and reagent kit
A cauliflower leaf, qualitative detection technology, applied in the direction of microbial measurement/testing, biochemical equipment and methods, etc., can solve the problems of not expressing protein or unstable expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0076] Preparation of purified water
[0077] Distilled with tap water once, purified by a Millipore MILLI-Q PF PLUS water purifier, collected water with a resistivity ≥ 18.0MΩ.cm, and stored in a sterile bottle.
[0078] Preparation of positive control substance
[0079] Take the corresponding positive sample, and extract the DNA with the CTAB method sample processing reagent, and amplify the above-mentioned prepared DNA with a primer longer than the detection reaction amplification fragment (using dTTP instead of dUTP), using Promega's Wizard PCRPreps DNA Purification System kit was used to purify the target nucleic acid fragments in the above PCR products, and then 1.5% agarose gel electrophoresis was used to roughly quantify the purified amplification products, and then the target fragments were purified by Promega’s PGEM-TEasy Vector System II kit. Connect with the T vector; then transform the connected product into JM109 competent cells, and then use X-Gal and IPTG agar...
Embodiment 1
[0133] Select genetically modified and non-genetically modified agricultural products such as soybeans, corn, rape, tomatoes, and potatoes, and extract genomic DNA with the Promega magnetic bead method, specifically: weigh 100 mg of plant material and place it in a 2ml centrifuge tube; add 500ul LysisBuffer A and 5ul RNase A, mix with the material; add 250ul Lysis Buffer B, mix well, and let stand at room temperature for 10 minutes; add 750ul precipitation buffer, mix well and centrifuge at high speed (13000×g) for 10 minutes; draw the supernatant into a clean 2ml centrifuge tube , add 50ul magnetic powder solution, and mix well; add 0.8 volume of isopropanol to the mixture, mix well, and let it stand at room temperature for 5 minutes; put the centrifuge tube on the magnetic rack for 1 minute, remove the clarified liquid; take out the centrifuge tube, and then add 250ul Lysis Buffer B, mix well and place on the magnetic rack for 1 minute, remove the clarified liquid; wash the m...
Embodiment 2
[0146] The 5' end of the above probe 35s7248FAMB was labeled with biotin, and the 3' end was not labeled; the 5' end of the primer 35s7345r was labeled with fluorescein. The probes were pre-coated on the ELISA plate with avidin.
[0147] The DNA in the sample was extracted according to conventional methods (CTAB method or magnetic bead method). Consists of 10×PCR buffer, dNTPs, Mg 2+ , Taq enzyme, upstream and downstream primers, H 2 O, and a DNA template to form a PCR reaction system for amplification.
[0148] After the amplification is completed, denature the amplified product into a single strand and add it to the enzyme-linked plate coated with the probe. If there is specific amplification, one strand of the amplified product will hybridize with the probe, wash the plate, and add Enzyme-linked fluorescein antibody, wash the plate, add color reagent, develop color for 10 minutes, stop color development, read the OD value on the microplate reader, and judge the negative ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com