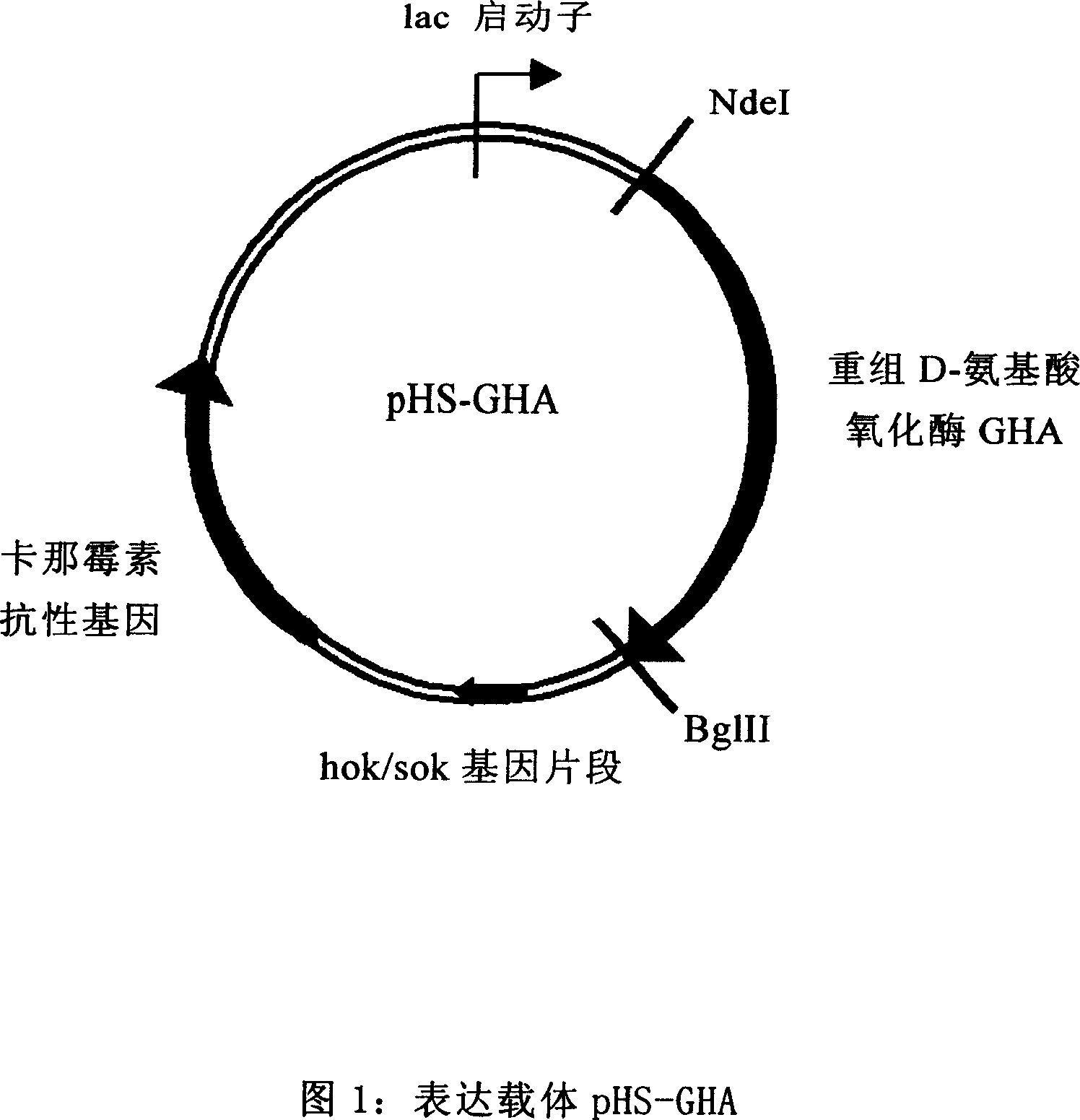
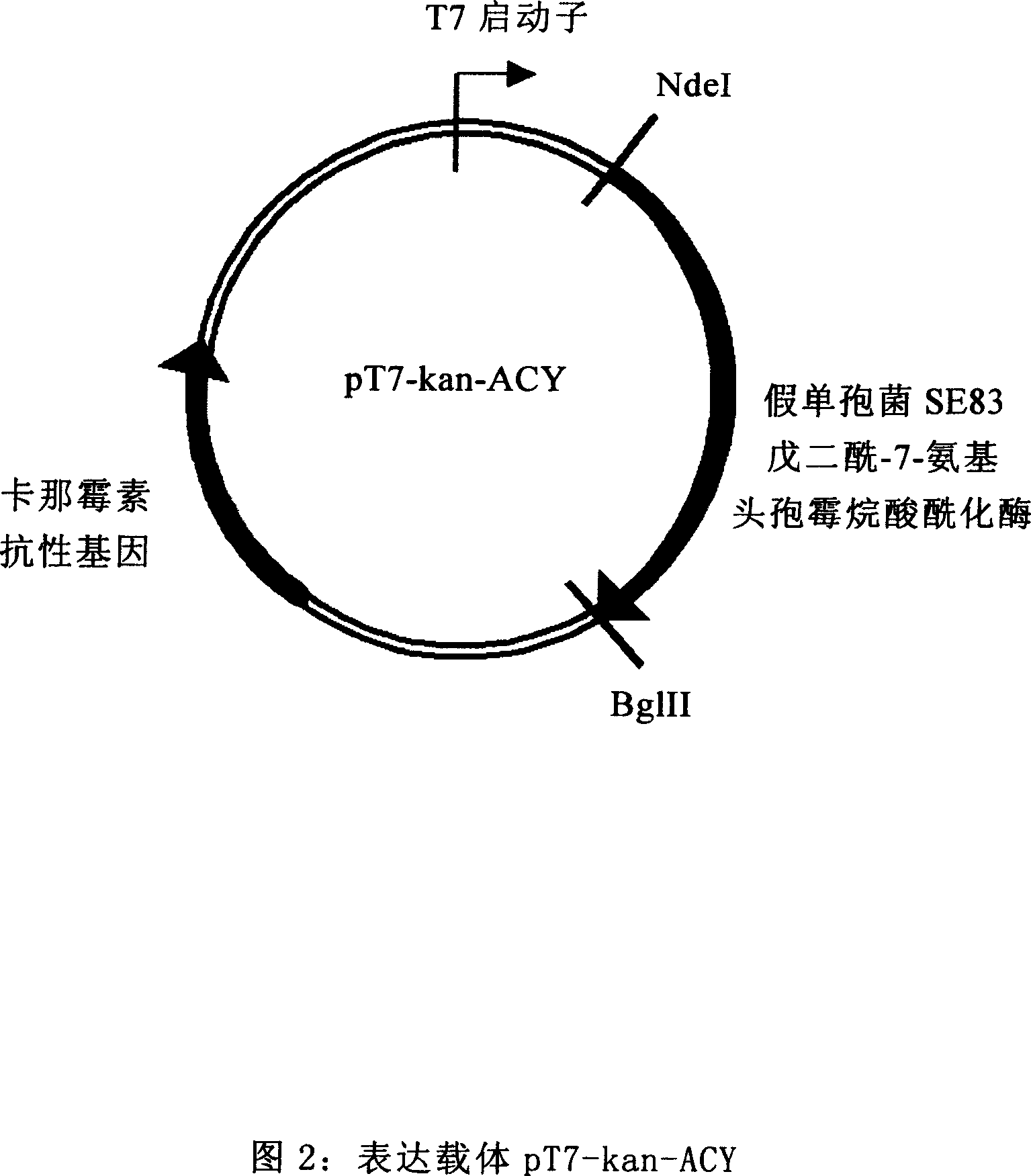
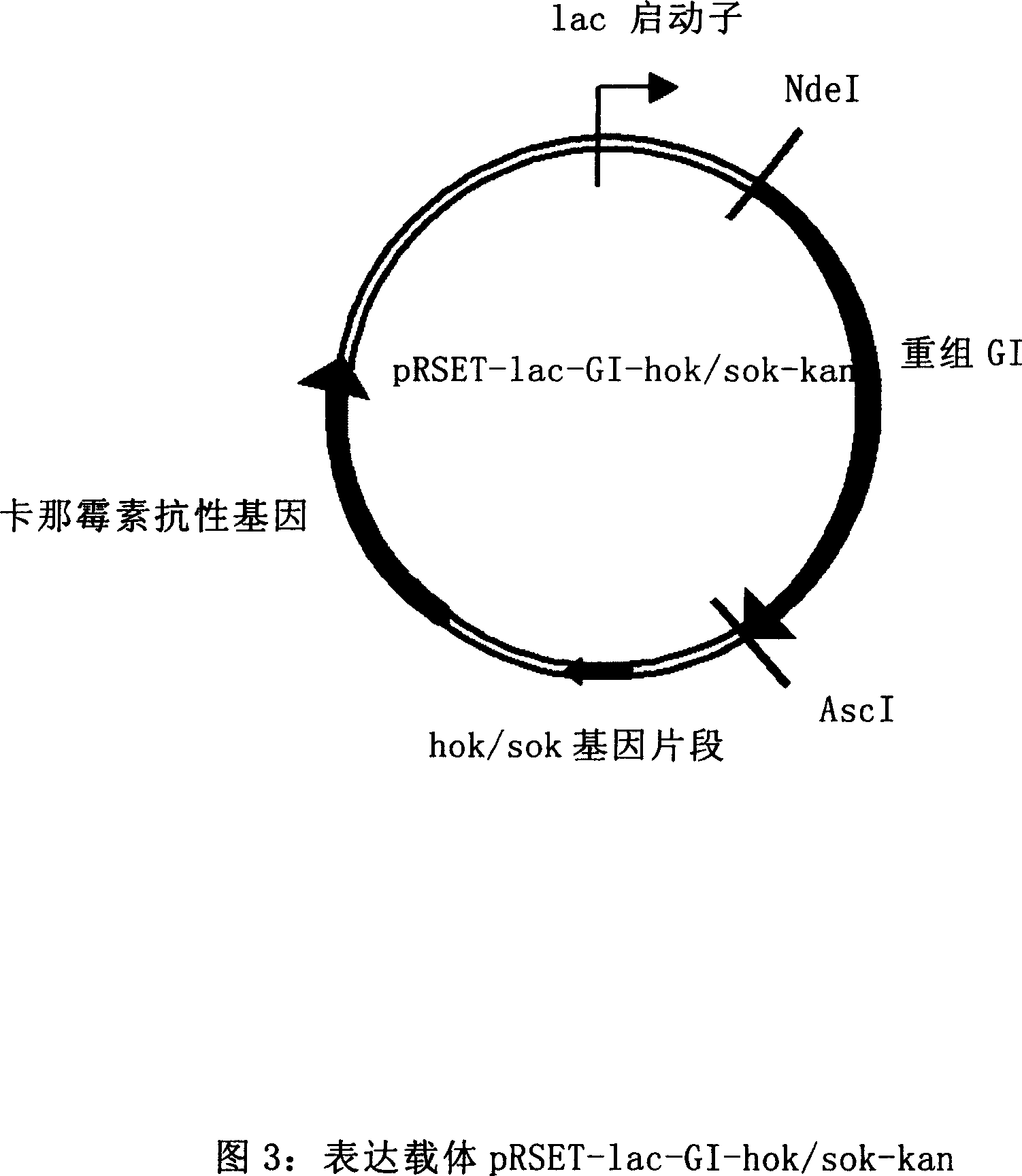
Colibacillus expression carrier and its application
An expression vector, a technology of Escherichia coli, applied in the field of bioengineering, can solve the problems of low yield and high cost of 7-aminocephalosporanic acid
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1: Construction of expression vector pRSET-kan
[0019] Design PCR primers according to the sequence of pRSET-A, specifically:
[0020] Upstream primer VET-F: 5’-CTGTCAGACCAAGTTTACTCATATATACTTTAG-3’;
[0021] Downstream primer VET-R: 5'-ACTCTTCCTTTTTCAATATTATTGAAGC-3'.
[0022] The PCR primers were designed according to the sequence of pET28b, specifically the upstream primer KAN-F: 5'-ATGAGCCATATTCAACGGGAAAC-3'; the downstream primer KAN-R: 5'-TTAGAAAAACTCATCGAGCATCAAATG-3'.
[0023] The PCR conditions for amplifying the pRSET-A fragment with the ampicillin resistance gene removed are: 50ng pRSET-A (Invitrogen), 0.4μM VET-F, 0.4μM VET-R, 50μM dATP, 50μM dTTP, 50μM dCTP, 50μM dGTP, 20mM Tris-HCl (pH8.8), 10mM KCl, 10mM (NH 4 ) 2 SO 4 , 2mM MgSO 4 , 0.1% Triton X-100, 2.5U Pfu DNA polymerase (Promega), adjust the reaction volume to 50 μL with sterile water. The PCR amplification reaction program is: 94°C, 5 minutes; 94°C, 1 minute, 50°C, 1 minute, 72°C, 4 minutes, cy...
Embodiment 2
[0026] Example 2: Construction of vector pRSET-lac-kan
[0027] Design PCR primers according to the sequence of pGEMT-Easy (Promega), specifically: upstream primer RBS-NdeI: 5’- CATATG TATATC TGTGTGAAATTG-3' (the NdeI restriction site is underlined, and the additional ribosome binding site is represented by the dashed line below); downstream primer RBS-AlwNI: 5’- CAGTGGCTG CTGCCAGTGGCGATAAGTC-3' (AlwNI restriction site is underlined).
[0028]Using pGEMT-Easy (Promega) as a template, PCR was performed with the above primers, and a 755bp product was amplified. The PCR conditions were: 50ng pGEMT-Easy (Promega), 0.4μMRBS-NdeI, 0.4μM RBS-AlwNI, 50μM dATP, 50μM dTTP, 50μM dCTP, 50μM dGTP, 20mM Tris-HCl (pH 8.8), 10mM KCl, 10mM (NH 4 ) 2 SO 4 , 2mM MgSO 4 , 0.1% Triton X-100, 2.5U Pfu DNA polymerase (Promega), adjust the reaction volume to 50 μL with sterile water. The PCR amplification reaction program is: 94°C, 5 minutes; 94°C, 1 minute, 50°C, 1 minute, 72°C, 4 minutes, cycle 35 t...
Embodiment 3
[0031] Example 3: Construction of vector pGEMT-Easy-GI
[0032] According to the known Thermoanaerobacterium saccharolyticum glucoseisomerase DNA sequence (GenBank L09699), design PCR primers, specifically: upstream primer (GI-NdeI):
[0033] 5’- CATATG AATAAATATTTTGAGAACGTATCTAAAATA-3' (NdeI restriction site is underlined);
[0034] Downstream primer (GI-EcoRI): 5’- GATATC TTAA TTATTCTGCAAAC-3' (EcoRI restriction site is underlined, AscI restriction site is double-underlined).
[0035] Using Thermoanaerobacterium saccharolyticum (purchased from ATCC, USA) DNA as a template, PCR was performed with the above primers, and a 1,336bp product was amplified. PCR conditions were: 50ng T.saccharolyticum DNA, 0.4μM GI-NdeI, 0.4μM GI-EcoRI, 50μM dATP, 50μM dTTP, 50μM dCTP, 50μM dGTP, 20mM Tris-HCl (pH 8.8), 10mM KCl, 10mM (NH 4 ) 2 SO 4 , 2mM MgSO 4 , 0.1% Triton X-100, 2.5U Platinum Taq High Fidelity DNA polymerase (Invitrogen), adjust the reaction volume to 50 μL with sterile water. Th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com