Cells and non-human organisms containing predetermined genomic modifications and positive-negative selection methods and vectors for making same
a technology of predetermined genomic modifications and positive-negative selection methods, applied in the field of cells and non-human organisms, can solve the problems of adverse effects of transgenic animals, “leakage” of transgene expression, and disruption of endogenous genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Inactivation at the Int-2 Locus in Mouse ES Cells
1. PNS Vector Construction
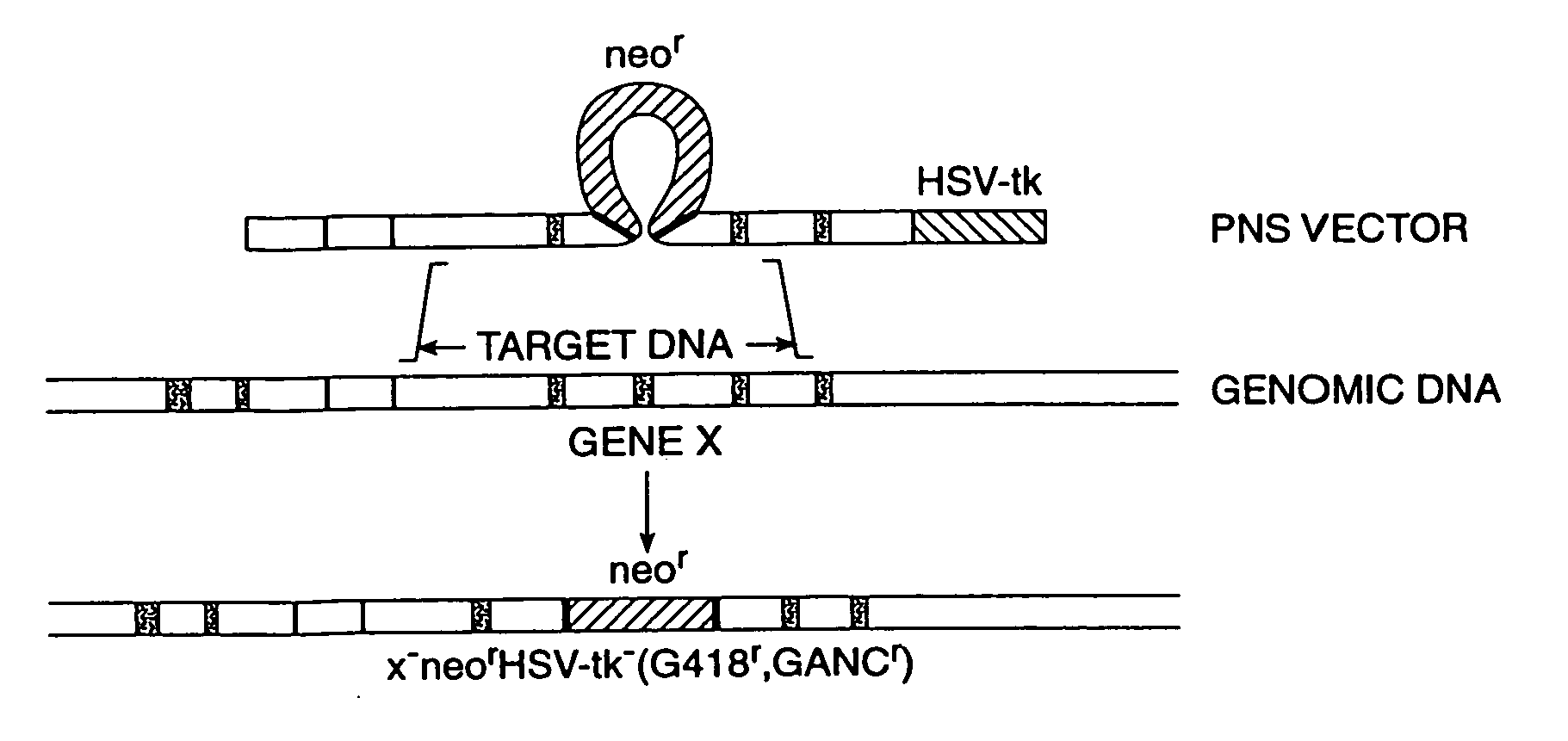
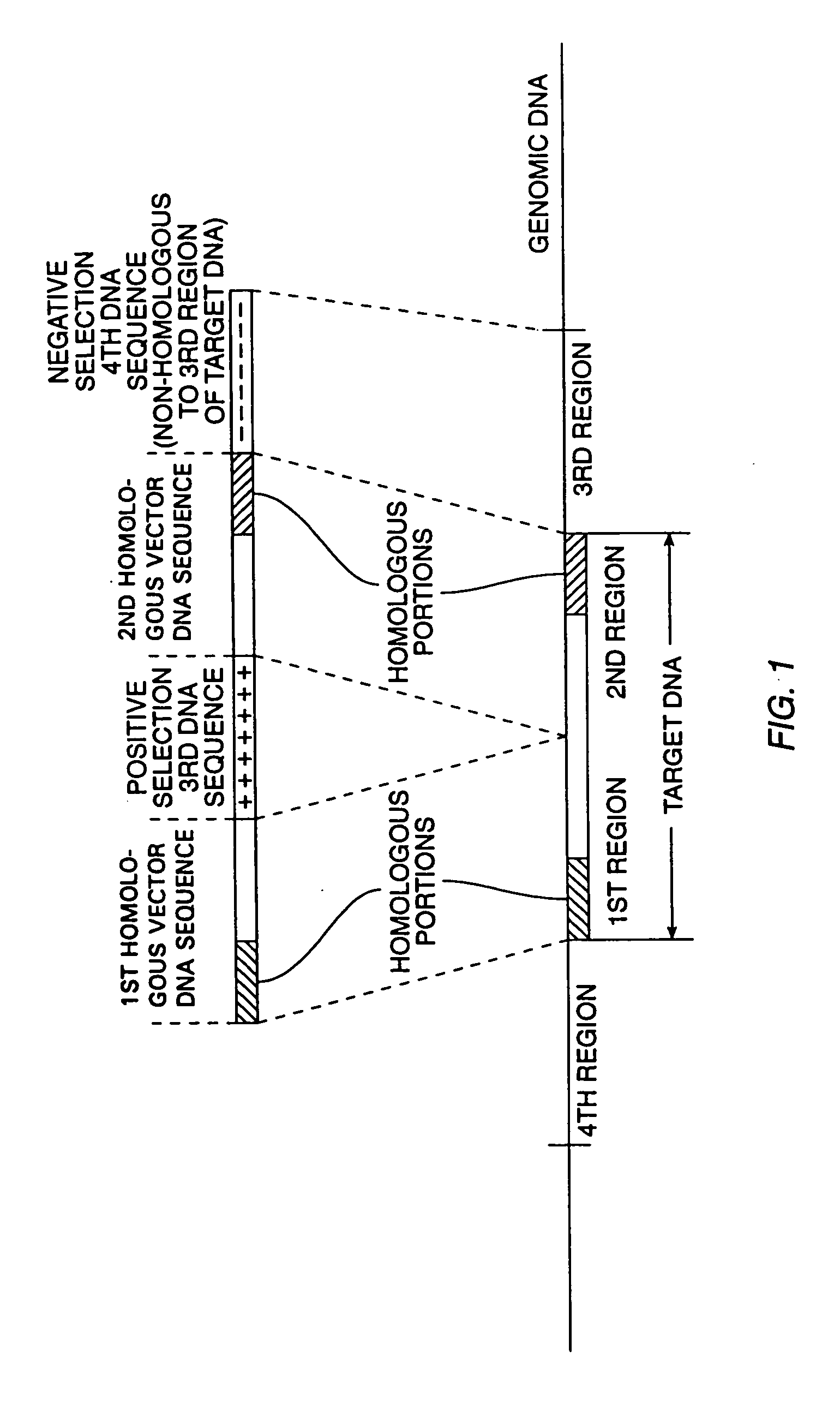
[0097] The PNS vector, pINT-2-N / TK, is described in Mansour, et al. (1988), Nature, 336, 349. This vector was used to disrupt the proto-oncogene, INT-2, in mouse ES cells. As shown in FIG. 5c, it contains DNA sequences 1 and 2 homologous to the target INT-2 genomic sequences in mouse ES cells. These homologous sequences were obtained from a plasmid referred to as pAT-153 (Peters, et al. (1983), Cell, 33, 369). DNA sequence 3, the positive selection moiety of the PNS vector was the Neo gene from the plasmid pMCINeo described in Thomas, et al. (1987), Cell, 51, 503; DNA sequence 4, the negative selection element of the vector, was the HSV-TK gene derived from the plasmid pIC-19-R / TK which is widely available in the scientific community.
[0098] Plasmid pIC19R / MC1-TK (FIG. 5d) contains the HSV-TK gene engineered for expression in ES cells (Mansour, et al. (1988), Nature, 336, 348-352). The TK gene, flanked by ...
example 2
Disruption at the hox1.4 Locus in Mouse ES Cells
[0108] Disruption of the hox1.4 locus was performed by methods similar to those described to disrupt the int-2 locus. There were two major differences between these two disruption strategies. First, the PNS vector, pHOX1.4N / TK-TK2 (FIG. 6), used to disrupt the hox1.4 locus contained two negative selection markers, i.e., a DNA sequence 5 encoding a second negative selection marker was included on the PNS vector at the end opposite to DNA sequence 4 encoding the first negative selection marker. DNA sequence 5 contained the tk gene isolate d from HSV-type 2. It functioned as a negative-selectable marker by the same method as the original HSV-tk gene, but the two tk genes are 20% non-homologous. This non-homology further inhibits r combination between DNA sequences 4 and 5 in the vector which might have inhibited gene-targeting. The second difference between the int-2 and the hox1.4 disruption strategies is that the vector pHOX1.4N / TK-TK2...
example 3
Inactivation of Other Hox Genes
[0111] The methods described in Examples 1 and 2 have also been used to disrupt the hox1.3, hox1.6, hox2.3, and int-1 loci in ES cells. The genomic sequences for each of these loci (isolated from the same-Dash library containing the hox1.4 clone) were used to construct PNS vectors to target disruption of these genes. All of these PNS vectors contain the Neo gene from pMCi-Neo as the positive selection marker and the HSV-tk and HSV-tk2 sequences as negative selection markers.
TABLE VOther Murine Developmental Genes Inactivated by PNSNeo-InsertionLocusGenomic FragmentSequence Ref.Sitehox1.311 kb Xba-HindIIITournier-Iasserve,EcoRI-site inet al. (1989),homeo-domainMCE, 9, 2273hox1.613 kb partial RIBaron, et al. (1987),BglII-site inEMBO, 6, 2977homeo-domainhox2.312 kb BamHIHart, et al. (1987),BglII-site inGenomics, 1, 182homeo-domainint-113 kb BglIIvan Ooyen et al.XhoI-site in(1984), Cell, 39, 233exon 2
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| resistance | aaaaa | aaaaa |
| absolute frequency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com