Efficient generation of adenovirus-based libraries by positive selection of adenoviral recombinants through ectopic expression of the adenovirus protease
a technology of protease and adenoviral recombinant, which is applied in the field of generating adenoviral recombinant vectors and adenoviral recombinant expression libraries, can solve the problems of virus replication incompetence, inability to multiply in infected host cells, and trigger selective destruction of targeted cells, etc., to achieve the effect of reducing the number of adenoviral recombinants, reducing the number of adenoviral
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
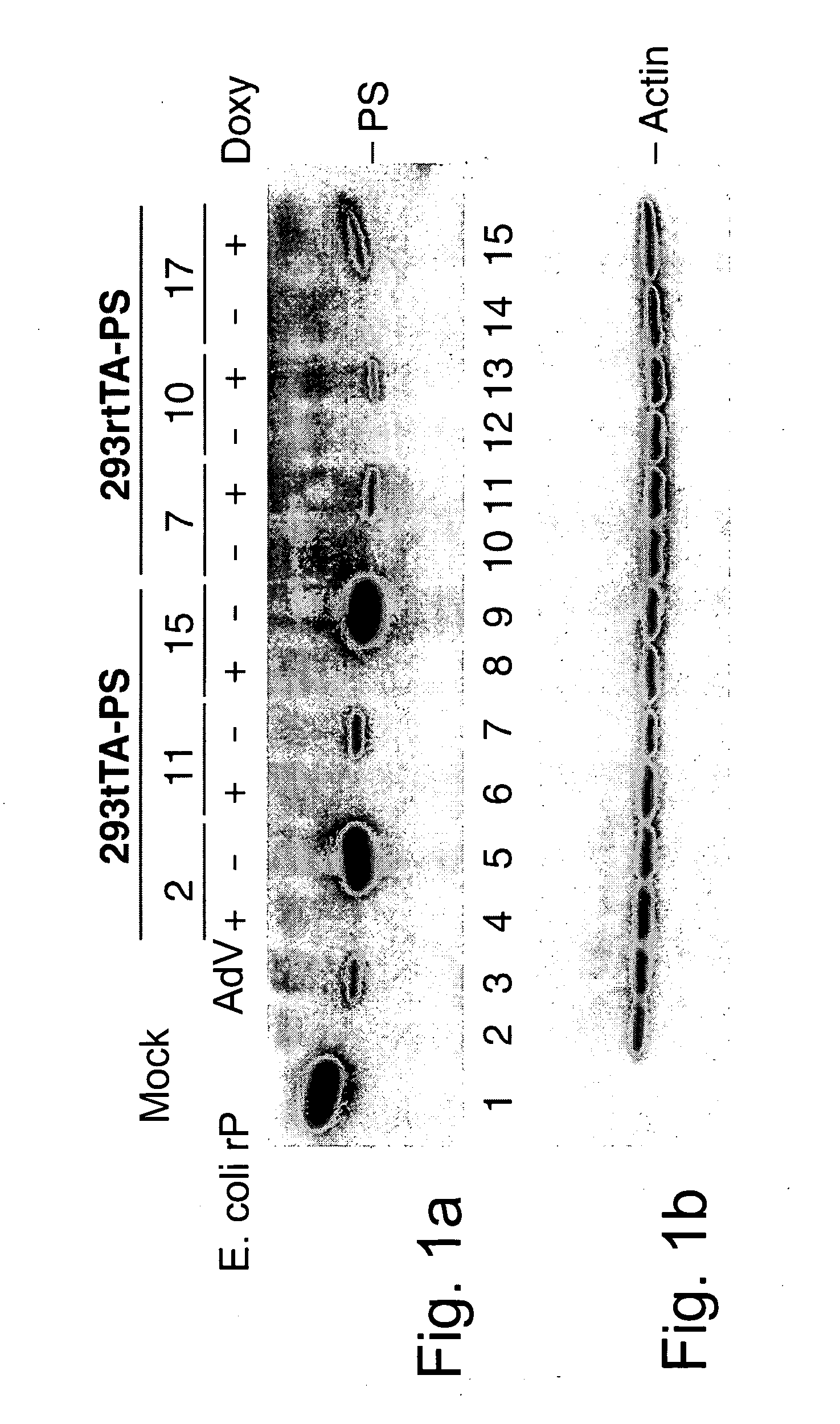
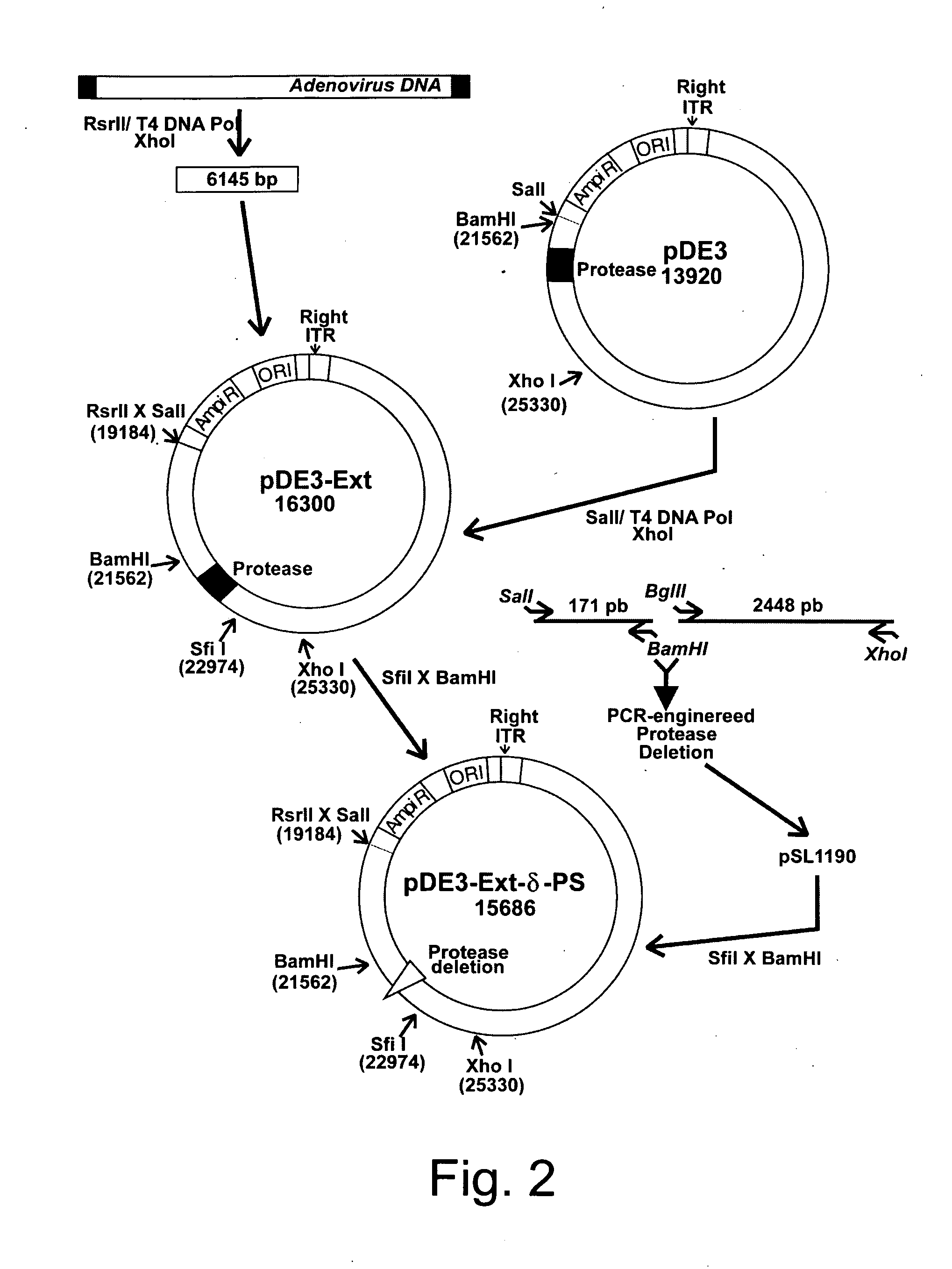
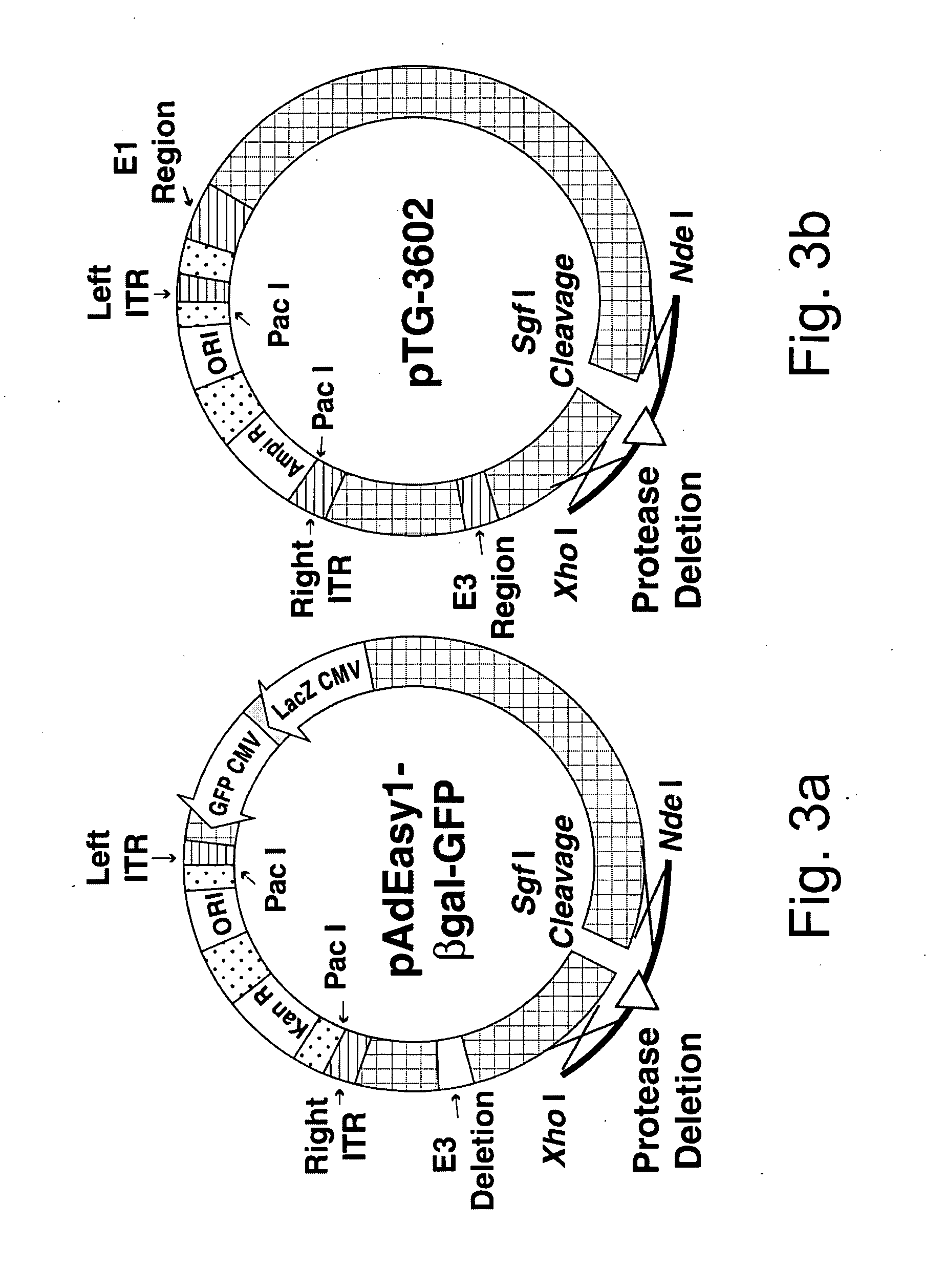
Generation and Isolation of 293 Cell Lines Transformed with Ad2 Protease Gene
[0070] Cell lines were generated by co-transfection and selection with appropriate agents as summarized in Table 1.
TABLE 1Analysis of the clones obtained from transformationof 293 cells with proteaseSelectedPlasmids usedSelectionClonesClonesPositiveCellsfor transfectionagentobtainedanalyzedclones293pTR5 / PS-DC / GFP+G418>50177tTApTKNeo293pTR5 / PS-DC / GFP+hygro->50149rtTAp3′SSmycin
[0071] 293 tTA cells were co-transfected with pTR5 / PS-DC / GFP and pTKNeo, while 293 rtTA were co-transfected with the same plasmid and p3'SS. After a 48 hour recovery, transfected cells were submitted to a three weeks selection by either G418 (500 μg / ml) for 293 tTA or hygromycin (150 μg / ml) for 293 rtTA. During this time, fresh medium and drug were applied to cells twice a week. Throughout the selection process, GFP expression was monitored on aliquots by flow cytometry analysis. Cells were then sorted using the multiwell automated ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com