Nonnucleoside inhibitors of reverse transcriptase, composite binding pocket and methods for use thereof
a reverse transcriptase and non-nucleoside technology, applied in the field of non-nucleoside inhibitors of reverse transcriptase, can solve the problems of limited crystal structure of rt nni complex, inability to predict favorable binding of apa in the tnk binding site, and limited application of rt-tnk complex analysis, etc., to achieve potent anti-hiv activity, inhibit rt activity, and high selectivity index
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Modeling Procedure
Construction of the Composite NNI Binding Pocket
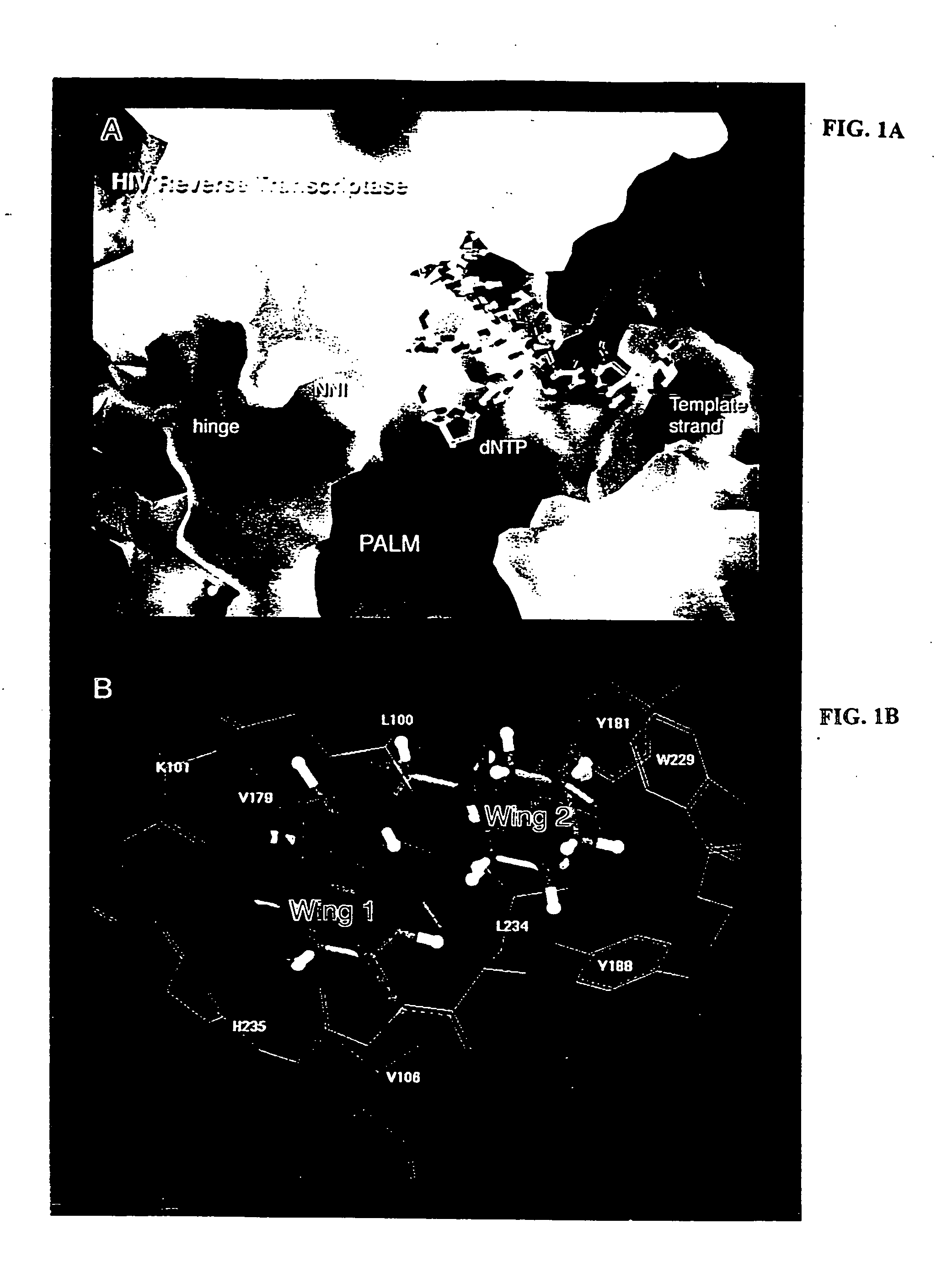
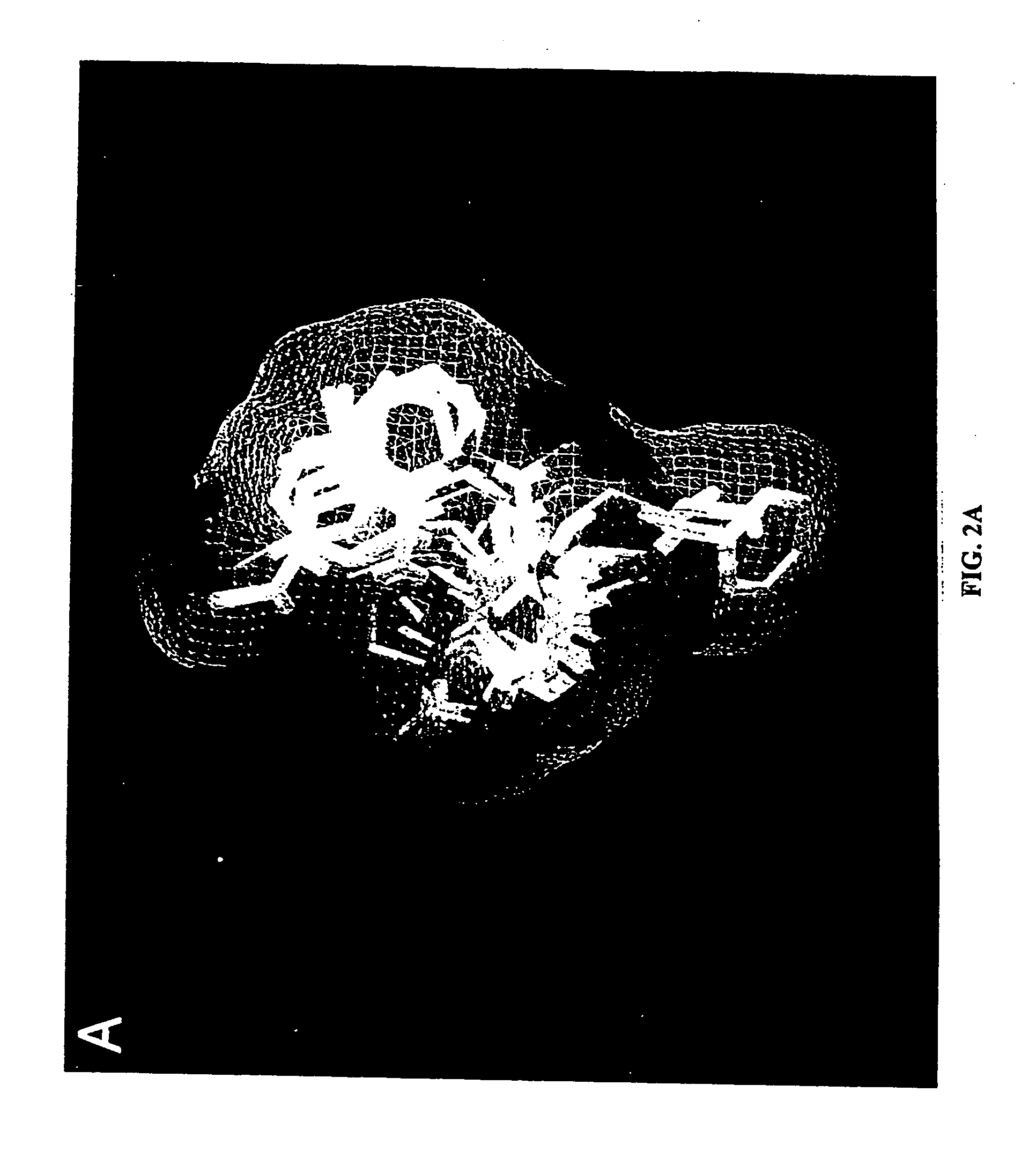
[0137] A novel model of the NNI binding pocket of RT was constructed by superimposing nine individual RT-NNI crystal structures and then generating a van der Waals surface which encompassed all of the overlaid ligands. This “composite binding pocket” surprisingly reveals a different and unexpectedly larger NNI binding site than shown in or predictable from any of the individual structures and serves as a probe to more accurately define the potentially usable space in the binding site (FIG. 2A).
[0138] Modeling studies were based on the construction of a binding pocket which encompassed the superimposed crystal structure coordinates of all known RT-NNI complexes, including nine different structures of RT complexed with HEPT, MKC, TNK, APA, Nevirapine, N-ethyl Nevirapine derivative, 9-Cl TIBO (Ren, J. et al., Structure, 1995, 3, 915-926); 9-Cl TIBO (Das, K. et al., J. Mol. Biol., 1996, 264, 1085-1100) and 8-Cl-TIBO (...
example 2
Predicted Efficacy of HEPT Derivatives
[0159] Compounds listed in Table 1 have been modeled into the NNI binding site of RT (RT / MKC 422 complex) using the docking procedure. The modeled positions were compared with the composite binding pocket of the invention, having the coordinates set forth in Table 9. Modeling was followed by analysis with the LUDI score function.
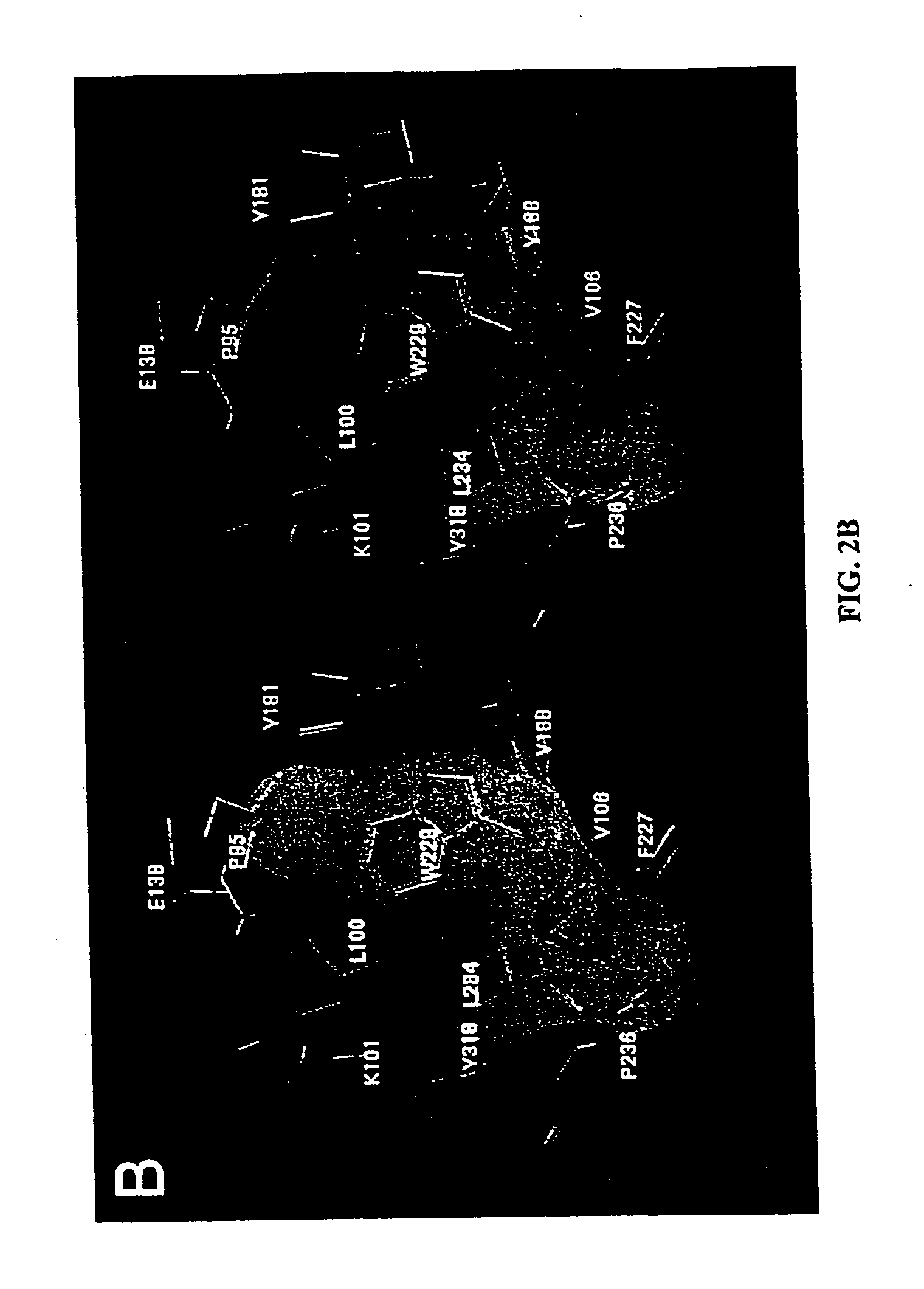
[0160] All of the positions of the compounds with top scores fall into the butterfly-shaped binding site, with the benzyl ring residing in wing 1 and the thymine ring in the wing 2 (FIG. 2). For all compounds tested, the benzyl ring is near Trp229 and the N−1 group is near Pro236, a typical position observed in crystal structures (FIG. 1B). The trend of calculated values listed in Table 1 shows that the Ki value decreases as a result of three factors: para substituents (R2) removed from the benzyl ring, larger alkyl groups added to the thymine ring (R1), and sulfur atoms substituted for oxygen (at X and / or Y). The mode...
example 3
DABO Derivatives
[0163] All chemicals were used as received from Aldrich Chemical Company (Milwaukee, Wis.). All reactions were carried out under nitrogen. Column chromatography was performed using EM Science silica gel 60 and one of the following solvents: ethyl acetate, methanol, chloroform, hexane, or methylene chloride. Nuclear magnetic resonance (NMR) spectra were recorded on a Varian (Palo Alto, Calif.) 300 MHz instrument (Mercury 2000 model) and chemical shifts (δ) are reported in parts per million (ppm) relative to tetramethylsilane as an internal standard at 0 ppm. 13C NMR spectra were recorded at 75 MHz in CDCl3 on the same instrument using a proton decoupling technique. The chemical shifts reported for 13C NMR are referenced to the chloroform triplet at 77 ppm. Melting points were measured using a Mel-Temp 3.0 (Laboratory Devices Inc., Holliston, Mass.) melting apparatus and are uncorrected. UV spectra were recorded from a Beckmann (Fullerton, Calif.) ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| distance | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| body weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com