Cytoplasmic inhibition of gene expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of Tomato Mosaic Virus cDNA
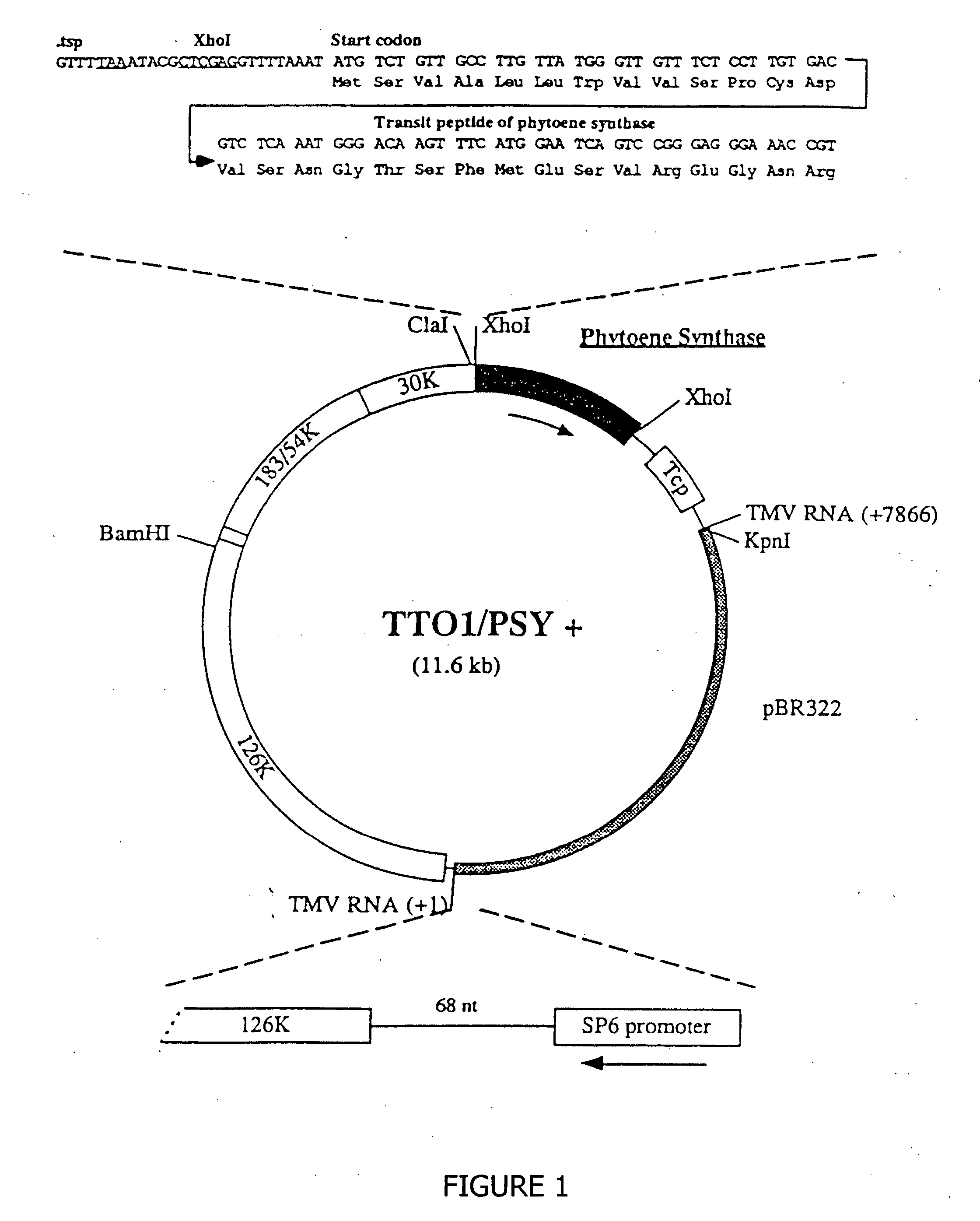
[0035] An 861 bp fragment (5524-6384) from the tomato mosaic virus (fruit necrosis strain F; ToMV-F) containing the putative coat protein subgenomic promoter, coat protein gene, and the 3′ end was isolated by PCR using ToMV primers 5′ CTCGCAAAGTTTCGAACCAAATCCTC 3′ (SEQ ID NO: 1) (upstream) and 540 CGGGGTACCTGGGCCCCAACCGGGGGTTCCGGGGG3′ (SEQ ID NO: 2) (downstream) and subcloned into the HincII site of pBluescript KS-. A hybrid virus consisting of TMV-U1 and ToMV-F was constructed by swapping an 874-bp XhoI-KpnI ToMV fragment into pBGC152 (Kumagai, et al., Proc. Natl. Acad. Sci. USA 90:427-430 (1993)), creating plasmid TTO1. The inserted fragment was verified by dideoxynucleotide sequencing. A unique AvrII site was inserted downstream of the XhoI site in TTO1 by PCR mutagenesis, creating plasmid TTO1A, using the following oligonucleotides:
(SEQ ID NO: 3)5′ TCCTCGAGCCTAGGCTCGCAAAGTTTCGAACCAAATCCTCA 3′(upstream),(SEQ ID NO: 2)5′ CGGGGTACCTGGGCCCCAAC...
example 2
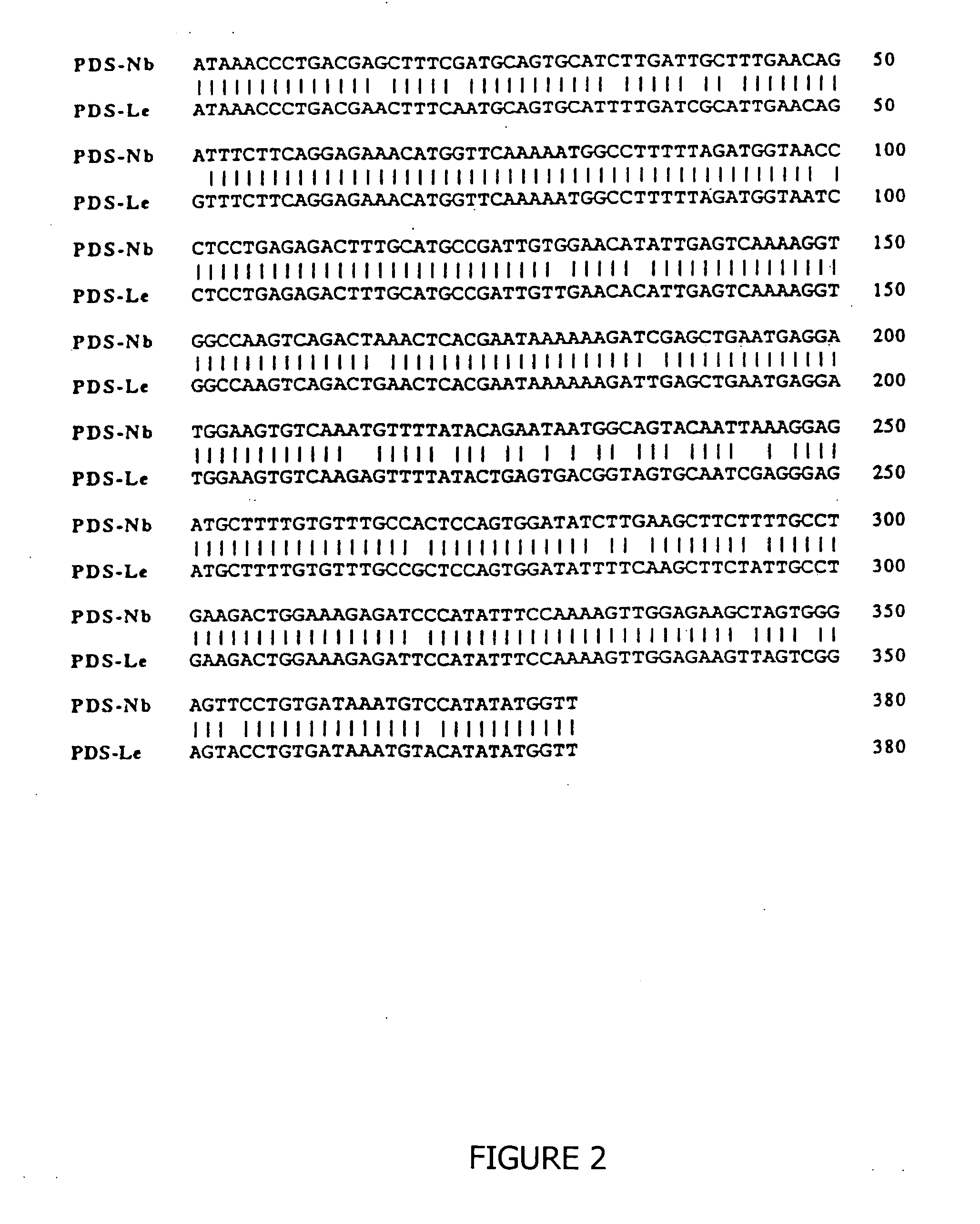
Isolation of a cDNA Encoding Tomato Phytoene Synthase and a Partial cDNA Encoding Tomato Phytoene Desaturase
[0036] Partial cDNAs were isolated from ripening tomato fruit RNA by polymerase chain reaction (PCR) using the following oligonucleotides: PSY, 5′ TATGTATGGTGCAGAAGAACAGAT 3′ (SEQ ID NO:4) (upstream), 5′ AGTCGACTCTTCCTCTTCTGGCATC 3′ (SEQ ID NO: 5) (downstream); PDS, 5′ TGCTCGAGTGTGTTCTTCAGTTTTCTGTCA 3′ (SEQ ID NO: 6) (upstream), 5′ AACTCGAGCGCTTTGATTTCTCCGAAGCTT 3′ (SEQ ID NO: 7) (downstream). Approximately 3×104 colonies from a Lycopersicon esculentum cDNA library were screened by colony hybridization using a 32 P labelled tomato phytoene synthase PCR product. Hybridization was carried out at 42° C. for 48 h in 50% formamide, 5×SSC, 0.02 M phosphate buffer, 5× Denhart's solution, and 0.1 mg / ml sheared calf thymus DNA. Filters were washed at 65° C. in 0.1×SSC, 0.1% SDS prior to autoradiography. PCR products and the phyoene synthase cDNA clones were verified by dideoxynucleoti...
example 3
DNA Sequencing and Computer Analysis
[0037] A 1.2 Kb PstI, BamHI fragment containing the phytoene synthase cDNA and a 0.7 Kb the partial phytoene desaturase cDNA was subcloned into pBluescript KS+ (Stratagene, La Jolla, Calif.). The nucleotide sequencing of KS+ / PDS #38 and KS+ / 5′3′PSY was carried out by dideoxy termination using single stranded templates. Nucleotide sequence analysis and amino acid sequence comparisons were performed using PCGENE and DNA Inspector IIE programs.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Gene expression profile | aaaaa | aaaaa |
| Inhibition | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com