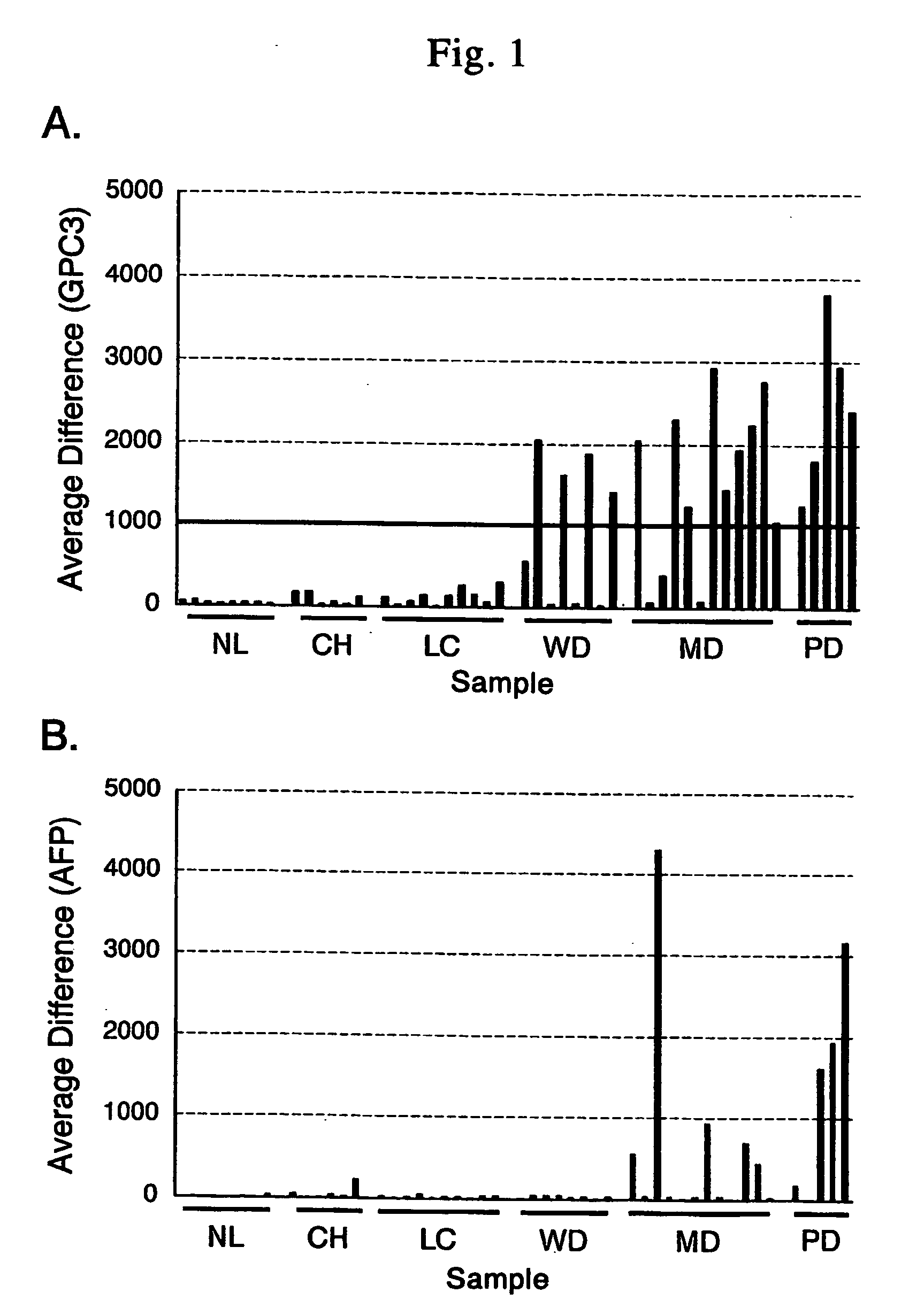
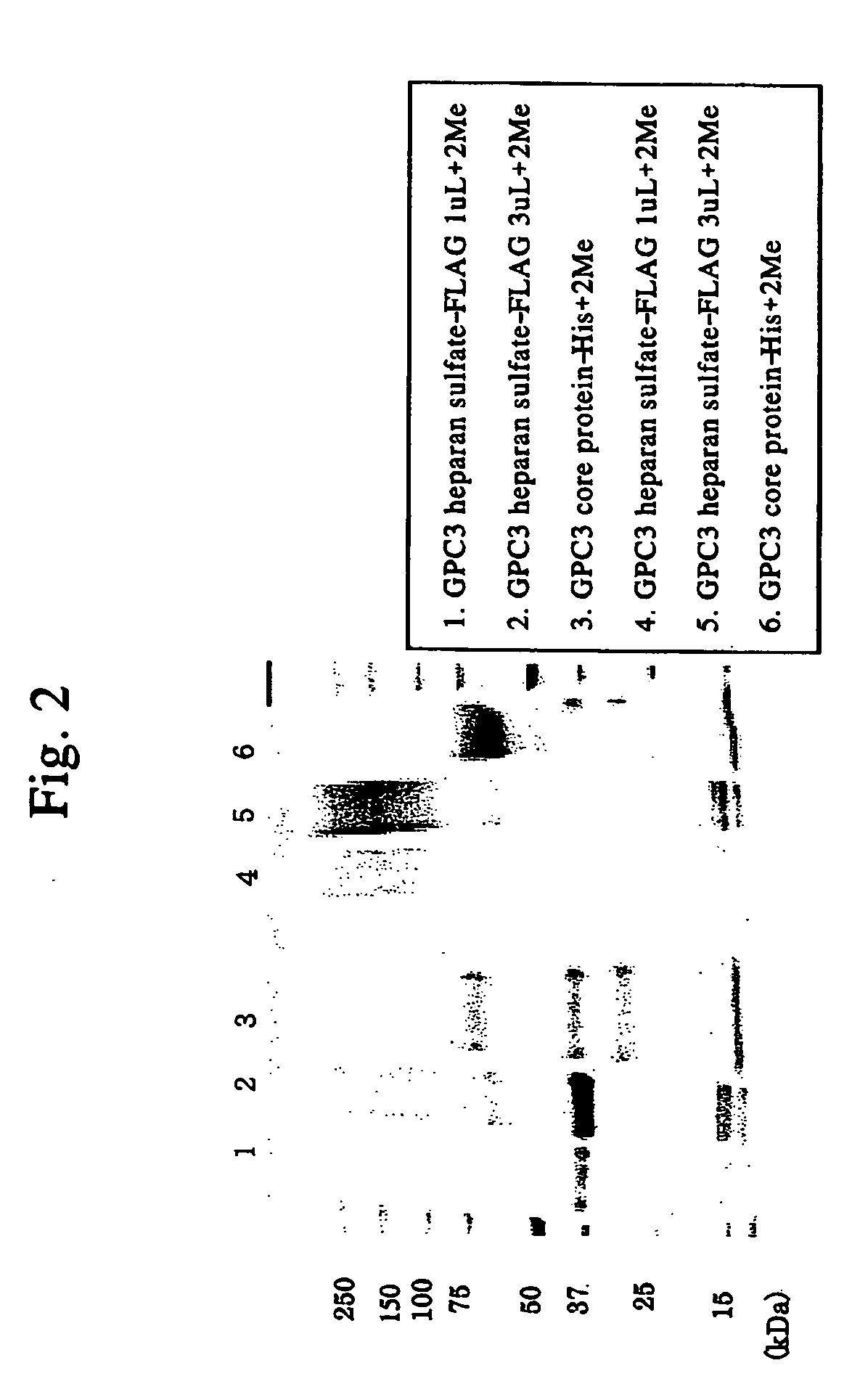
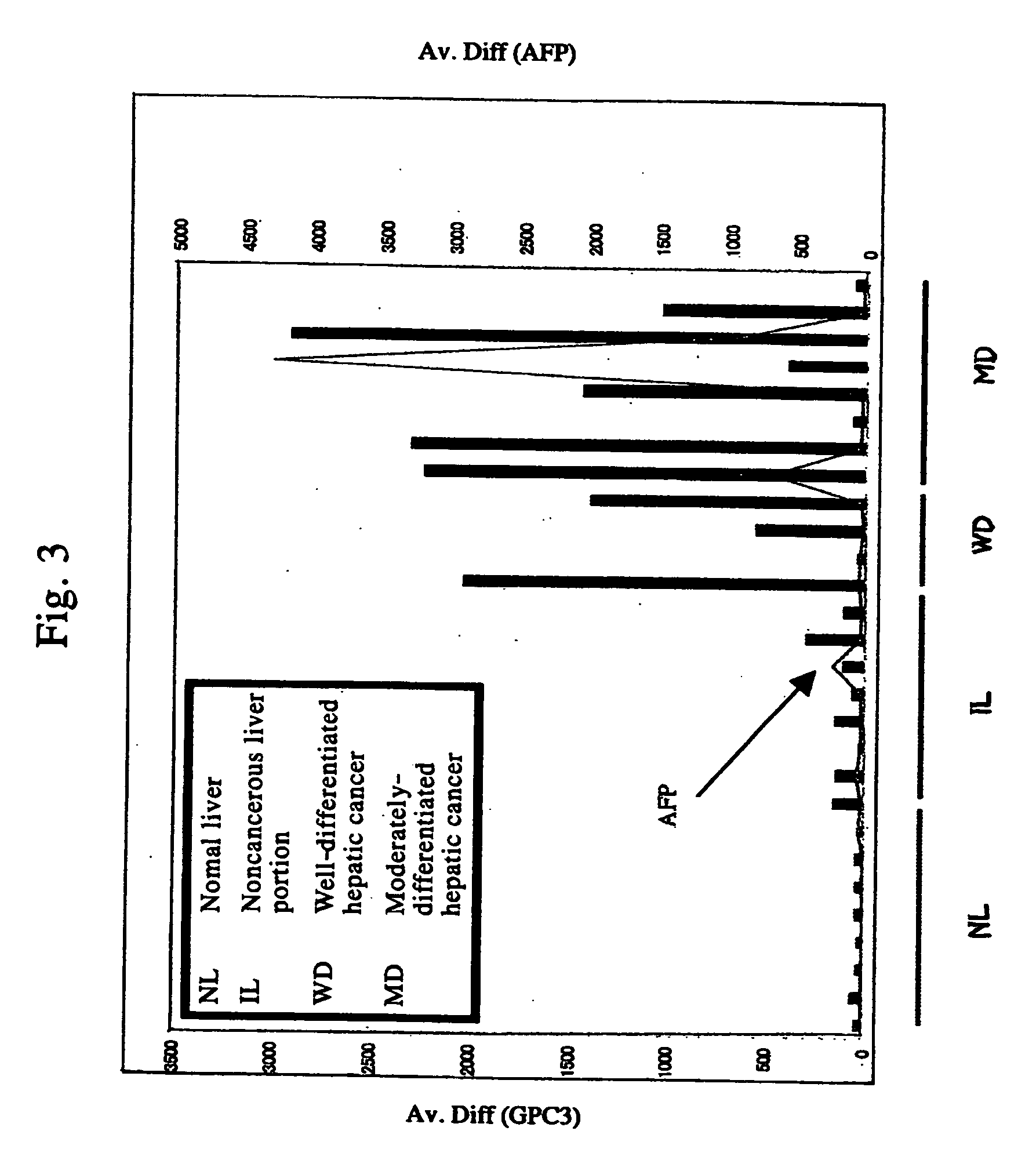
Method for diagnosing cancer by detecting gpc3
a cancer and soluble technology, applied in the field of soluble cancer markers in blood, can solve the problem of difficult detection of soluble gpc3, and achieve the effect of improving the stability of antibodies
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning and Expression Analysis of Human GPC (GPC3) cDNA
[0116] Cloning of the Full-Length cDNA Encoding Human Glypican 3 (Hereinafter Referred to as GPC3)
[0117] The full-length cDNA encoding human GPC3 was amplified by the PCR reaction using the 1st strand cDNA prepared by the standard method from colon cancer cell line Caco2 as a template, and an Advantage 2 kit (CLONTECH, Cat. No 8430-1). Specifically, 50 μl of a reaction solution containing 2 μl of the cDNA derived from Caco2, 1 μl of a sense primer (SEQ ID NO: 1), 1 μl of an antisense primer (SEQ ID NO: 2), 5 μl of Advantage2 10×PCR buffer, 8 μl of dNTP mix (1.25 mM), and 1.0 μl of Advantage polymerase mix was subjected to 35 cycles of a reaction cycle consisting of 94° C. for 1 minute, 63° C. for 30 seconds and 68° C. for 3 minutes. The amplified product (inserted in a TA vector pGEM-T easy using pGEM-T Easy Vector System I (Promega, Cat. No. A1360)) of the PCR was sequenced using an ABI3100 DNA sequencer to confirm that cDNA...
example 2
Preparation of Anti-GPC3 Antibodies
Preparation of Soluble Human GPC3
[0120] A soluble GPC3 protein lacking the hydrophobic region on the C-terminal side was prepared as a material for preparing anti-GPC3 antibodies.
[0121] A soluble GPC3 cDNA expression plasmid DNA was constructed using a plasmid DNA containing full-length human GPC3 cDNA provided by the Research Center for Advanced Science and Technology, The University of Tokyo. PCR was performed using a downstream primer (5′-ATA GAA TTC CAC CAT GGC CGG GAC CGT GCG C-3′ (SEQ ID NO: 5)) designed to eliminate the hydrophobic region (amino acid 564 to amino acid 580) on the C-terminal side and an upstream primer (5′-ATA GGA TCC CTT CAG CGG GGA ATG AAC GTT C-3′ (SEQ ID NO: 6)) containing an EcoR I recognition sequence and a Kozak sequence. The thus obtained PCR fragment (1711 bp) was cloned into pCXND2-Flag. The thus prepared expression plasmid DNA was introduced into a CHO cell line DXB11. A CHO cell line highly expressing soluble ...
example 3
Detection of Soluble GPC3
Mouse Xenograft Model
[0133] 3,000,000 HepG2 human hepatic cancer cells were grafted subcutaneously to the abdominal portion of 6-week-old female SCID mice (Fox CHASE C.B-17 / Icr-scid Jcl, CLEA Japan, Inc.) and nude mice (BALB / cA Jcl-nu, CLEA Japan, Inc.). 53 days later (when tumor mass had been sufficiently formed), the whole blood was collected via the posterior vena cava of HepG2-grafted SCID mice #1, 3 and 4. Plasma was prepared using a Nipro neotube (vacuum blood-collecting tube, NIPRO, NT-EA0205) in the presence of EDTA-2Na and aprotinin, and then stored at −20° C. by the day of measurement. In addition, the whole blood was collected from HepG2-grafted SCID mice #2 on day 62 after grafting of HepG2, and from HepG2-grafted nude mice #1 and 2 on day 66 after grafting through the posterior vena cava. As a control, plasma was also prepared in a similar procedure from normal SCID mice of the same age.
Sandwich ELISA
[0134] To detect soluble GPC3 in blood,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com