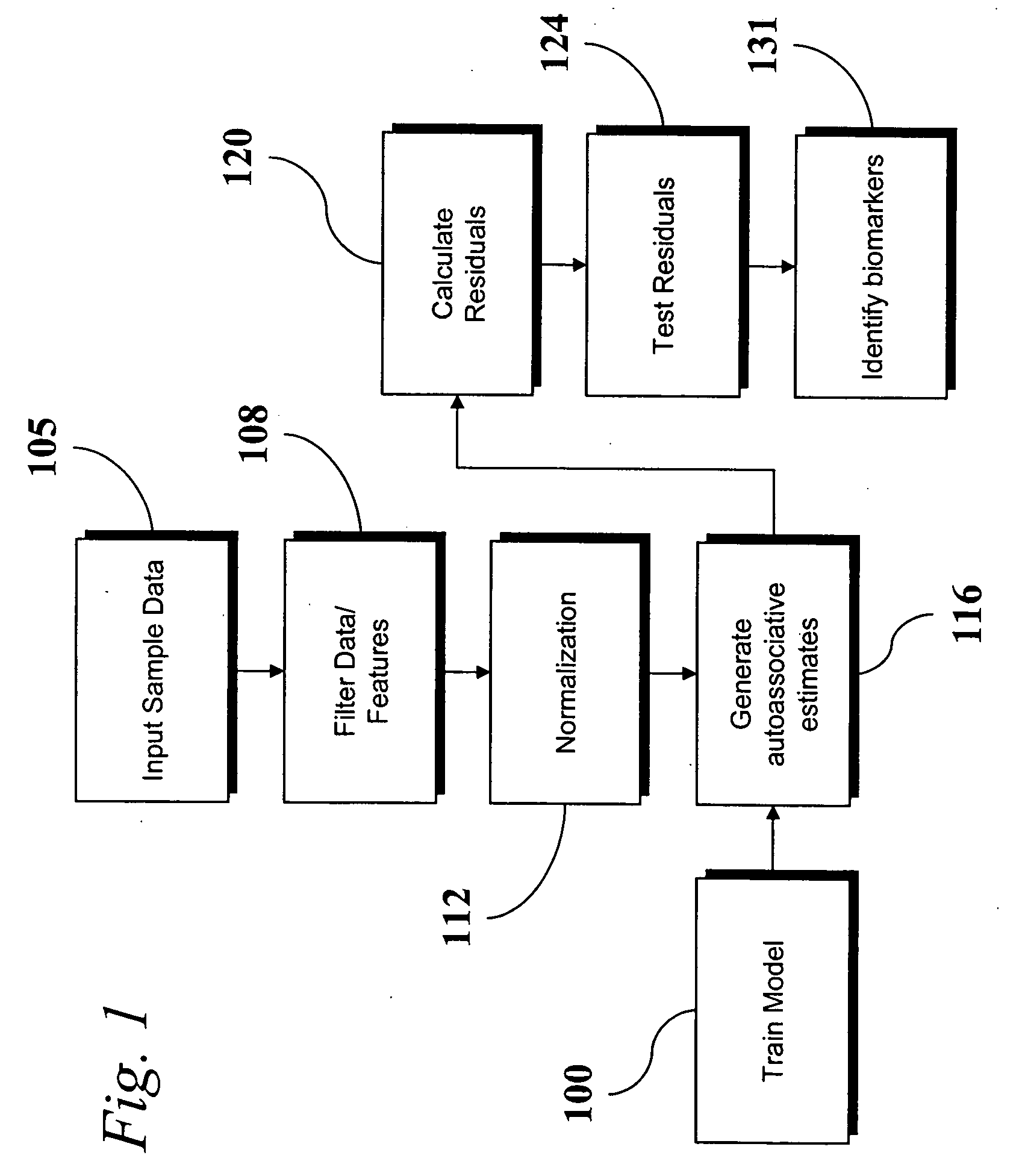
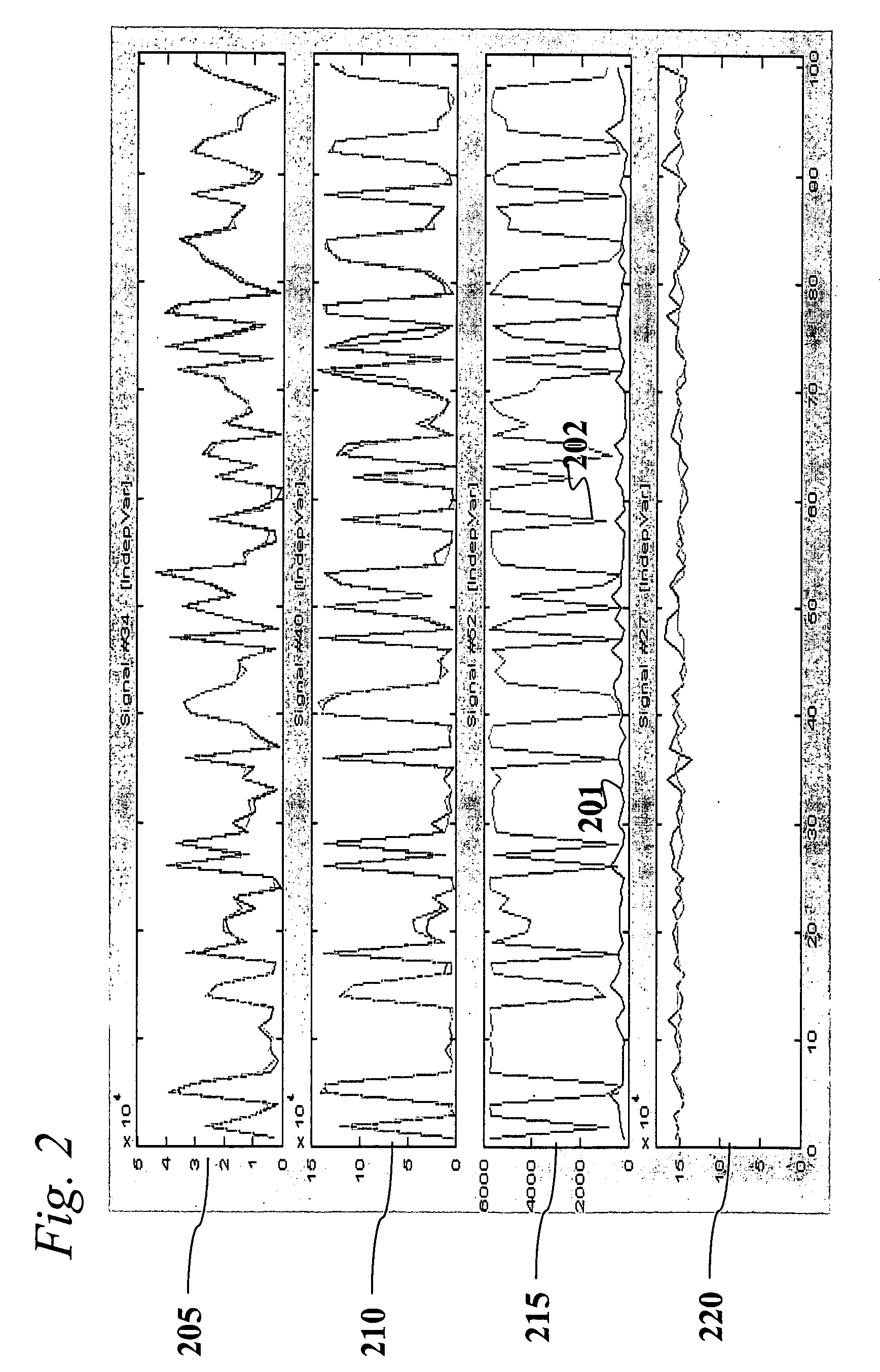
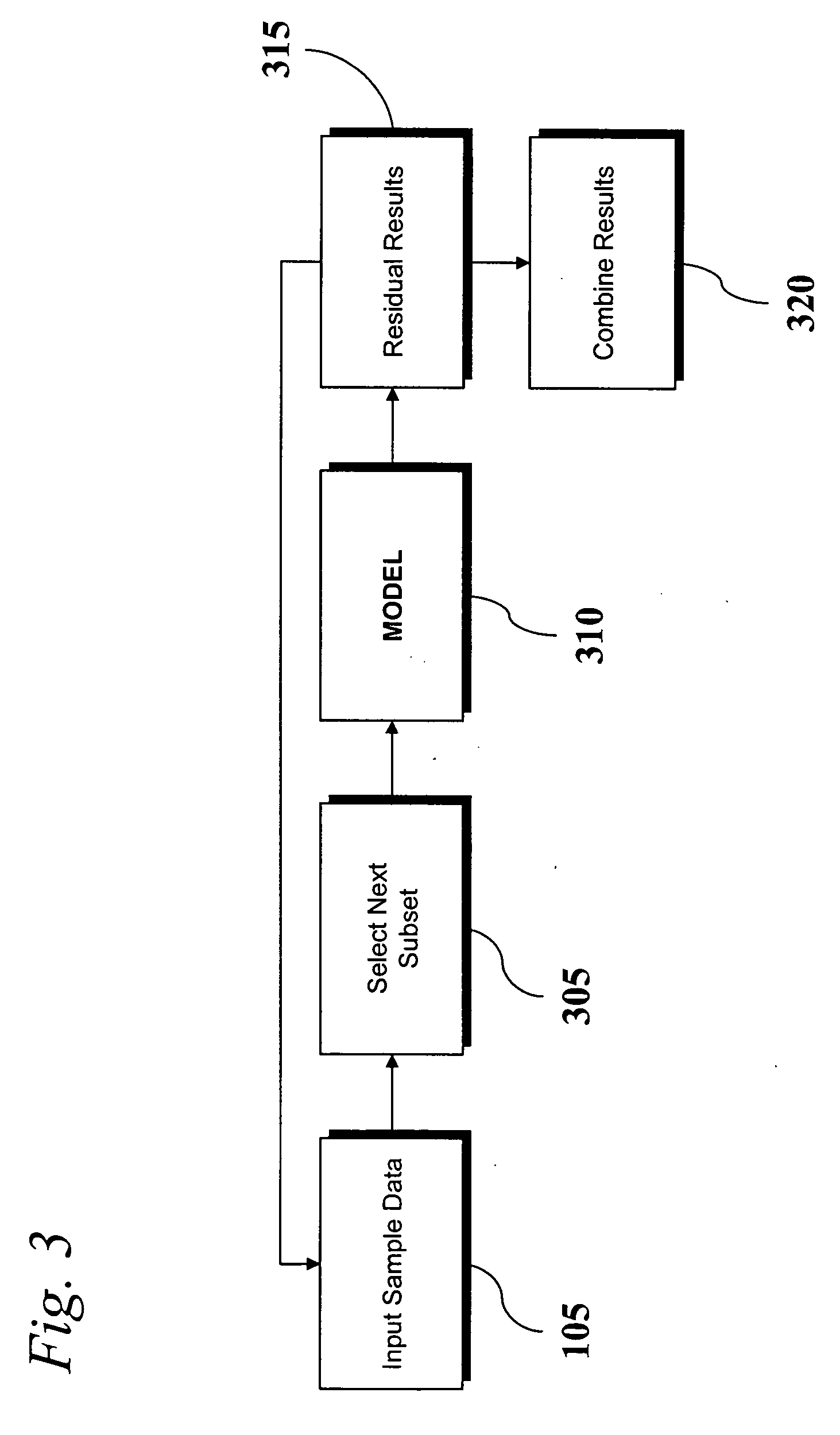
Analysis of transcriptomic data using similarity based modeling
a transcriptomic data and similarity-based modeling technology, applied in the field of transcriptomic data, can solve the problems of mis-regulation and upset of delicately balanced networks, and achieve the effects of reducing the complexity of the analysis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example
[0043] A simulated system comprising 15 constituents was developed whereby the 15 constituents related in their dynamic behavior to one another with varying degrees of linkage, emulating a regulatory network in a metabolic system. A set of reference data for this system, comprising observations of the 15 variables throughout various states of the dynamics of the system, was used to train a kernel-based model. Then, sets of normal and diseased observations, respectively, were generated, wherein one of the 15 constituents was perturbed to be slightly lower than it should be, regardless of its raw value. Turning to FIG. 10, a chart 1004 shows the value (quantity, expression level, etc.) for the suspect constituent, for the set of normal test observations. Each stem is a separate measurement of that constituent from the set of observations of “normal” specimens. Similarly is shown in chart 1005 the values for that same constituent in the set of observations of the “diseased” specimens. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com