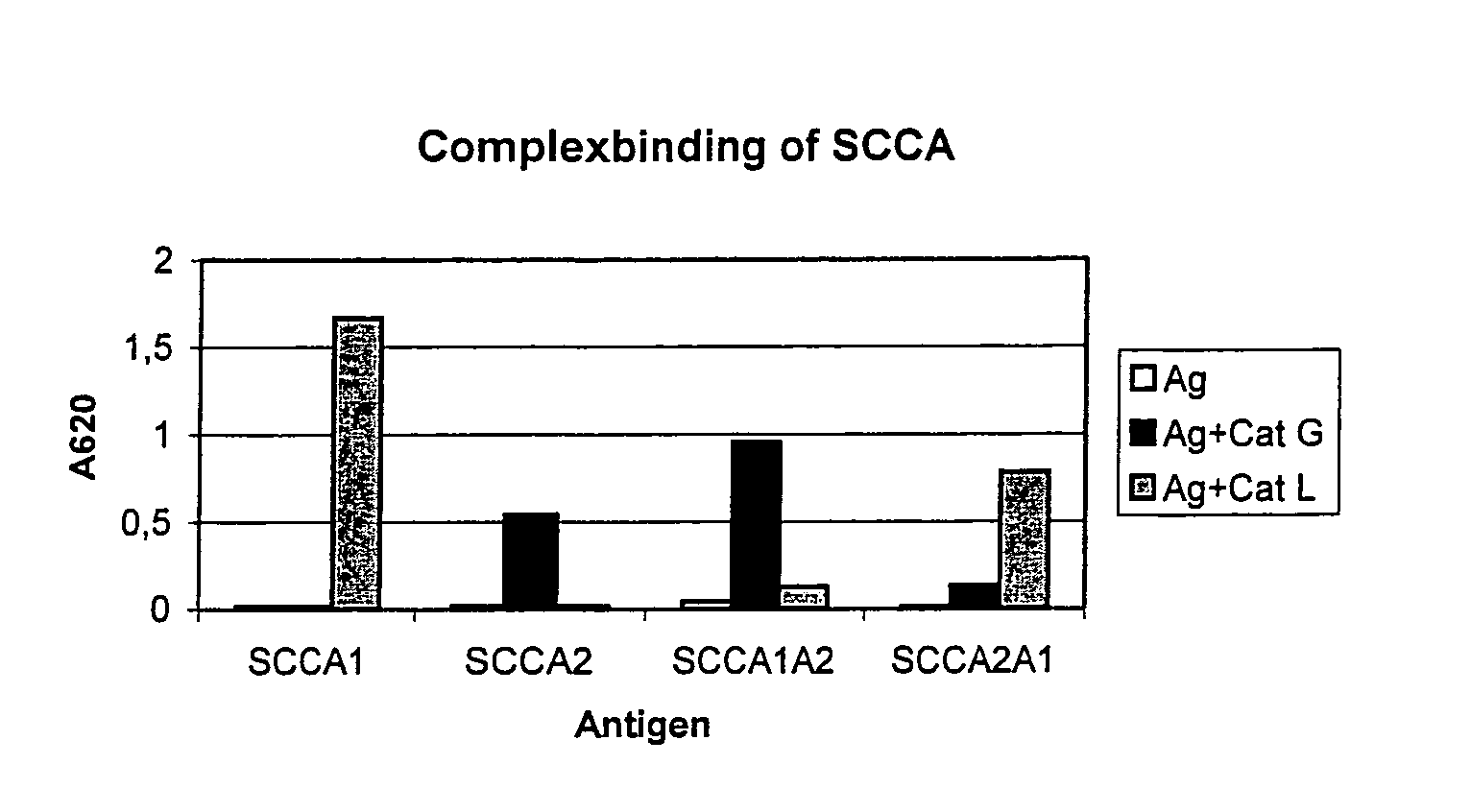
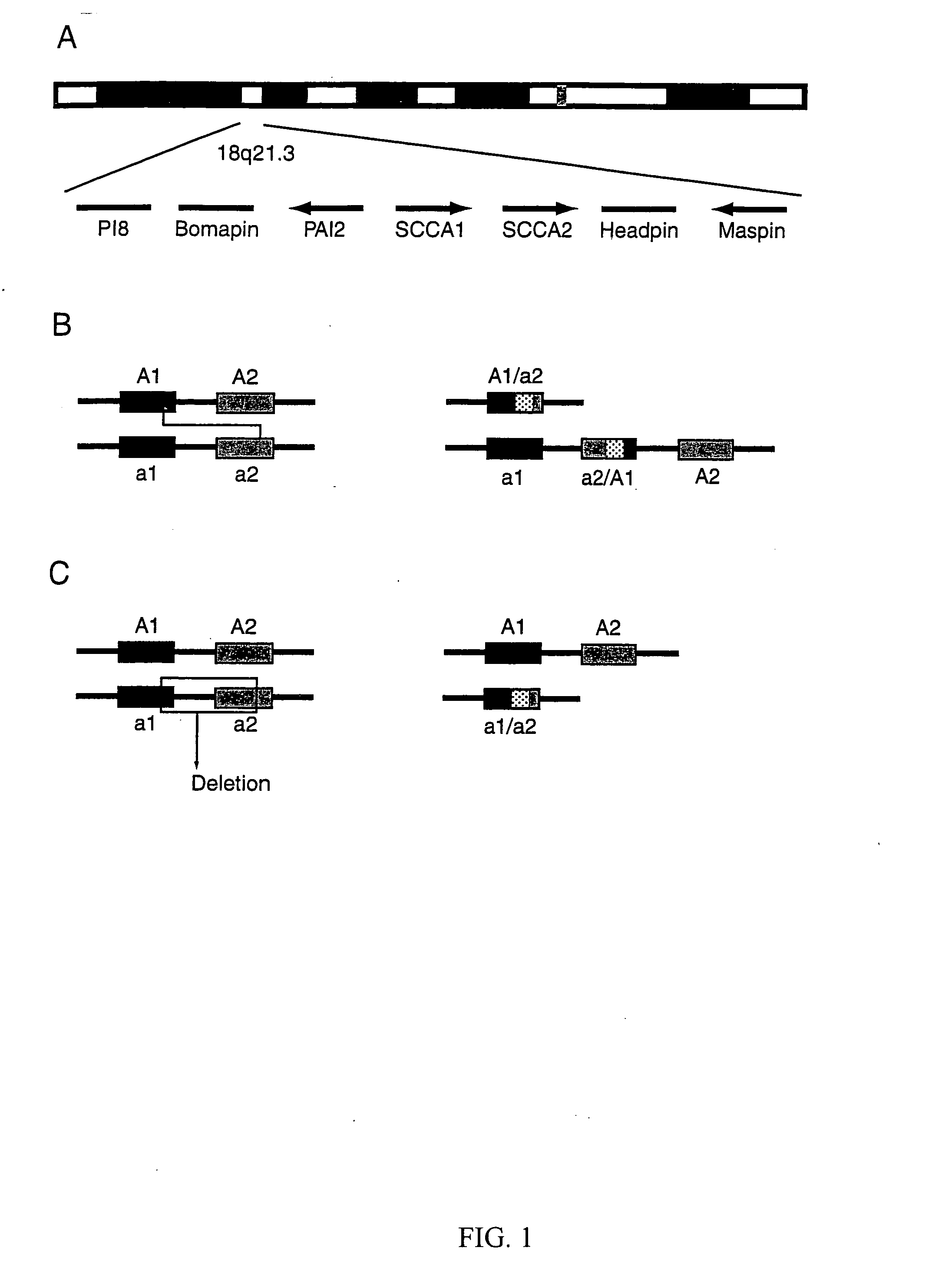Rearranged squamous cell carcinoma antigen genes II
a squamous cell carcinoma and antigen gene technology, applied in the field of fusion protein transcripts, can solve problems such as false positives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning of SCCA
1.1. PCR Amplification
[0021] mRNA from the cell-lines Caski (cervix), C4-I (cervix), A549 (lung), CaLu3 (ung), SkMes (lung), and RPMI2650 (pharynx) was prepared using QuickPrep Micro mRNA Purification kit (Pharmacia) and cDNA was prepared using First-Strand cDNA Synthesis kit (Pharmacia). A 1218 bp DNA fragment covering the coding sequence of SCCA was amplified by PCR in a 100 μl reaction containing 10 mM Tris-HCl pH 8.85, 25 mM KCl, 5 mM (NH4)2SO4, 2 mM MgSO4 (Boehringer), 0.2 mM DNTP (Pharmacia), 10 μM SCCA 1-7F (DNA sequences for all primers are shown in Table 1), 10 μM SCCA 391-397B, 2 μl cDNA and 2.5 U Pwo-polymerase (Boehringer). After denaturing samples for 5 min at 96° C. a total of 30 cycles were performed, each consisting of denaturation for 15 sec at 96° C., annealing for 15 sec at 60° C., and extension for 30 sec at 72° C. The PCR reaction was completed by a final extension for 10 min at 72° C.
TABLE 1PCR-primersPrimer nameSequence1. SCCA 1-7F5′-CGGGA...
example2
Protein Expression and Purification
2.1. Protein Expression
[0027] Expression conditions were determined by small-scale preparations. For large scale expression 500 ml cultures of 2×YT and 100 μg / ml ampicillin were inoculated with 5 ml over-night culture and grown at 37° C. Protein expression was induced at OD600=0.5-1.3 by adding IPTG to a final concentration of 0.1 mM. Cultures producing SCC1 were grown for 4-16 h, SCCA1 / A2 for 16-18 h. Cultures producing the SCCA2 protein were induced at OD600=1.2-1.4 and were grown for 2-3 h.
2.2. Protein Purification
[0028] Cells were harvested by centrifugation for 10 min at 2000 g, washed with 50 ml TE pH 8.0, and dissolved in 3 ml TE / g bacterial pellet. Lysozyme was added to a final concentration of 800 μg / g pellet and the mixtures were incubated on ice for 30-60 min and then frozen over night at −70° C. Magnesium chloride and DNase were added to a final concentration of 12 mM and 20 μg / g pellet, respectively. After incubation on ice for ...
example 3
DNA Analysis
3.1. Southern Blot Analysis
[0030] Approximately 10 μg of DNA prepared from SCC cell-lines as well as from blood samples from normal healthy volunteers, were digested with restriction endonucleases PstI or BamHI. Digested DNA were separated on 0.8% agarose and transferred to membranes (Hybond N+, Pharmacia). Filters were prehybridized for 1 h and hybridized over night at 60° C. in 20 ml of a solution containing 5×SSC, 0.1% SDS, 5% Dextrane sulfate, Liquid block (Pharmacia) diluted 1:20 and salmon sperm DNA 100 μg / ml. Probe concentration during hybridization was 10 ng / ml. After hybridization filters were stringency washed for 15 min in 1×SSC / 0.1% SDS and for 15 min in 0.2×SSC / 1% SDS, both at 60° C. Probe hybridization was detected using Gene Images CDP-Star detection module (Pharmacia) with minor modifications. Filters were blocked for 1 hour at room temperature in a solution containing liquid block diluted 1:7.5. Then they were incubated in buffer A (0.1M Tris, 0.3M N...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com